Fig. 2 |. Protein production and analyses of codon orthogonality in clonal SSOs.
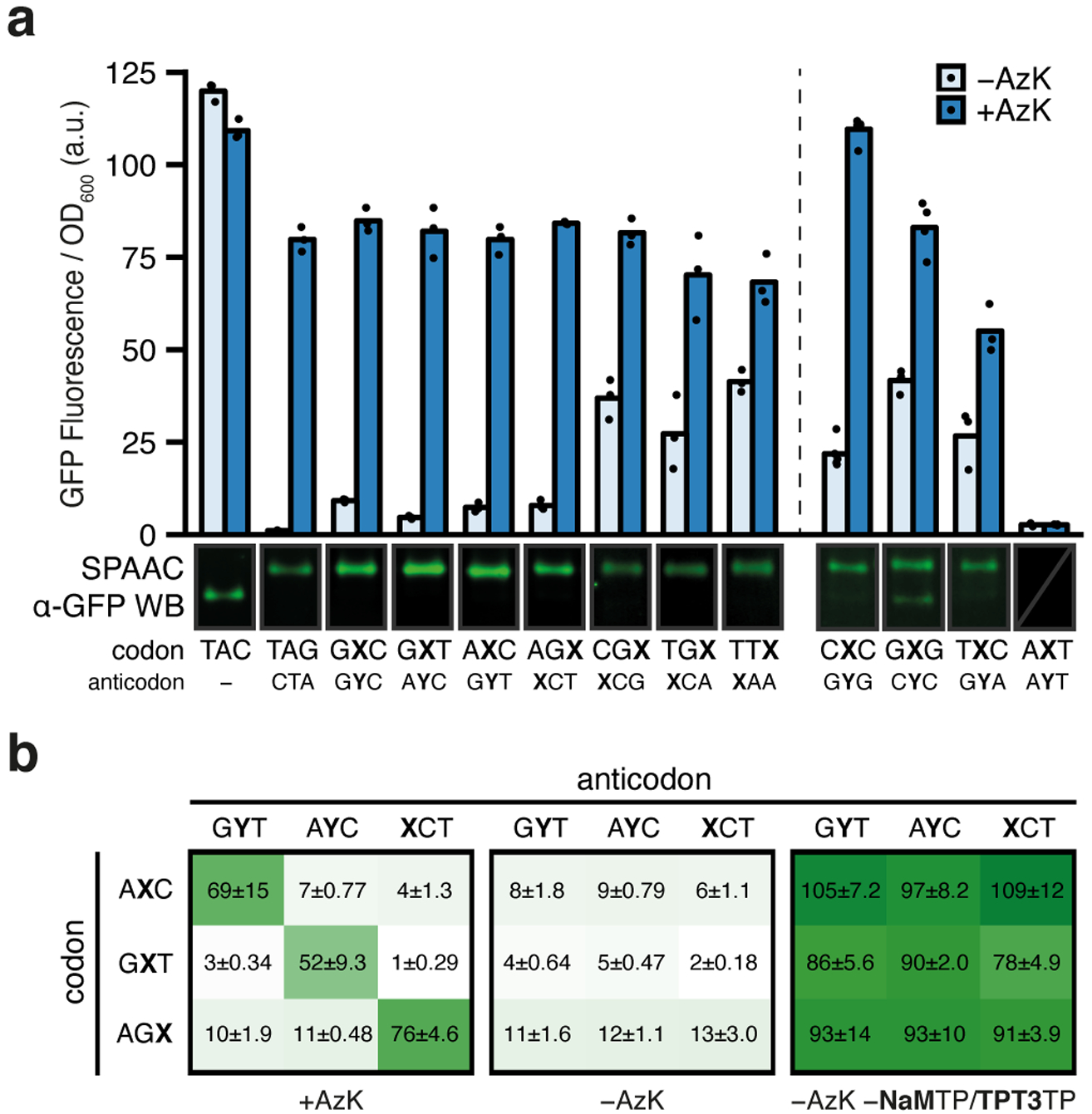
a, Normalized fluorescence from clonal SSOs at the endpoint of protein expression (i.e. t = 180 min after addition of aTc) for the seven top codons and anticodons (left) as well as the four other selected codons (right) both with and without AzK (left: n = 3, right: n = [5, 4, 3, 3]; biological replicates; mean with individual data points shown). The dashed line demarcates slight variations in how the clonal SSOs were prepared (see Methods). One representative cropped western blot of purified sfGFP, subjected to SPAAC with TAMRA-PEG4-DBCO from SSO cultures is shown (only α-GFP channel). All western blots shown in Supplementary Figs. 3 and 10. b, Normalized fluorescence from clonal SSO cultures at the endpoint of expression for all pairwise combinations of select codons and anticodons with and without AzK in media. Positive controls, without ribonucleoside triphosphates NaMTP and TPT3TP, which forces the incorporation of a natural ribonucleotide opposite the unnatural nucleotide in the template, were run to verify the integrity of the clone. Each culture was propagated from a single colony and mean ± standard deviation is indicated (black text; n = 3; biological replicates).
