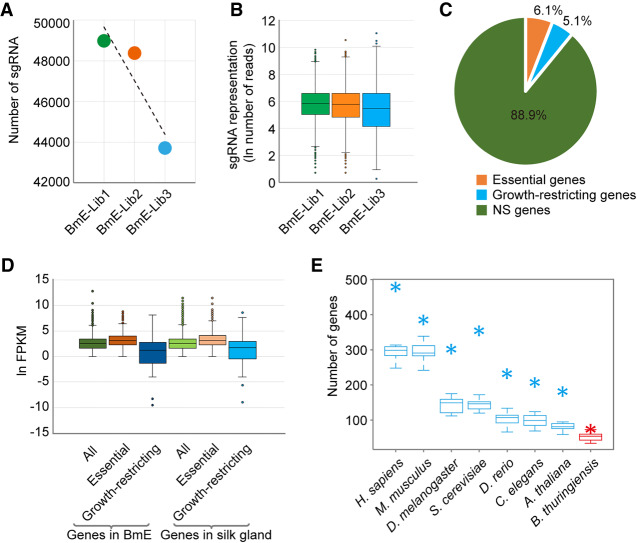
Figure 3.
Screening of essential and growth-restricting genes in BmE cells. (A,B) Changes in sgRNAs contained in BmEGCKLib over time. Approximately 4 × 107 cells of BmEGCKLib were harvested at each of three time points: immediately (BmE-Lib1), 1 mo (BmE-Lib2), and 2 mo (BmE-Lib3) after completion of zeocin selection. Overall the sgRNAs were gradually depleted but some of them were enriched over time. (C) Percentages of essential genes and growth-restricting genes in BmE cells. (D) Expression levels among all genes, essential genes, and growth-restricting genes in BmE cells and silk gland cells. (E) The overlap of essential genes among B. mori and diverse eukaryotic species (Homo sapiens, Mus musculus, Drosophila melanogaster, Saccharomyces cerevisiae, Danio rerio, Caenorhabditis elegans, and Arabidopsis thaliana), and a prokaryotic species (Bacillus thuringiensis). (Asterisks) The intersection of 1006 essential genes with B. mori orthologs of essential genes in the eight species; (box plots) the intersection of 1000 random genes with B. mori orthologs of essential genes in the eight species.