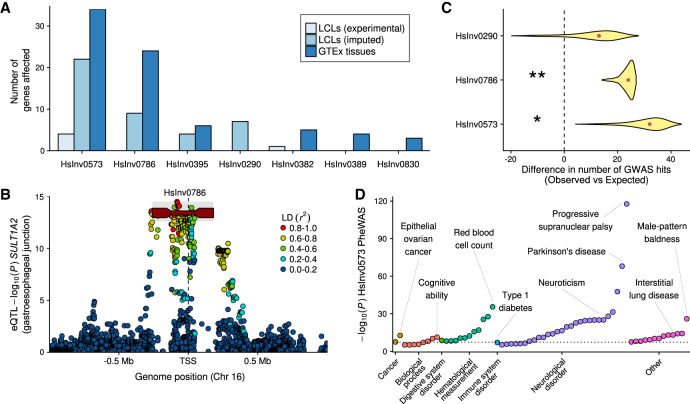
Figure 4.
Inversion effects on gene expression and phenotypic traits. (A) Number of significant associations at the gene level in each of the differential expression analyses (excluding the associations with specific transcripts). (B) Manhattan plot for cis-eQTL GTEx associations of gene SULT1A2 in gastroesophageal junction, showing inversion HsInv0786 (dark red line with rectangles representing the IRs) as potential lead variant. HsInv0786 eQTL P values and LD with neighboring variants (r2) were calculated by permuting samples with the same ancestry proportions as in GTEx samples (see Methods), and the P-value imputation range is shown in gray. (C) Enrichment of GWAS Catalog signals within individual inversions measured by the deviation in the observed minus expected value. Density distribution represents the 95% one-sided confidence interval with the median indicated by a red dot; one-tailed permutation test, (*) P < 0.05, (**) P < 0.01. (D) Phenome-wide association study (PheWAS) for inversion 17q21 (HsInv0573). Significant reported traits (P < 10−5) were grouped by GWAS Catalog ontology categories. Dotted line indicates genome-wide significance threshold (P = 5 × 10−8).