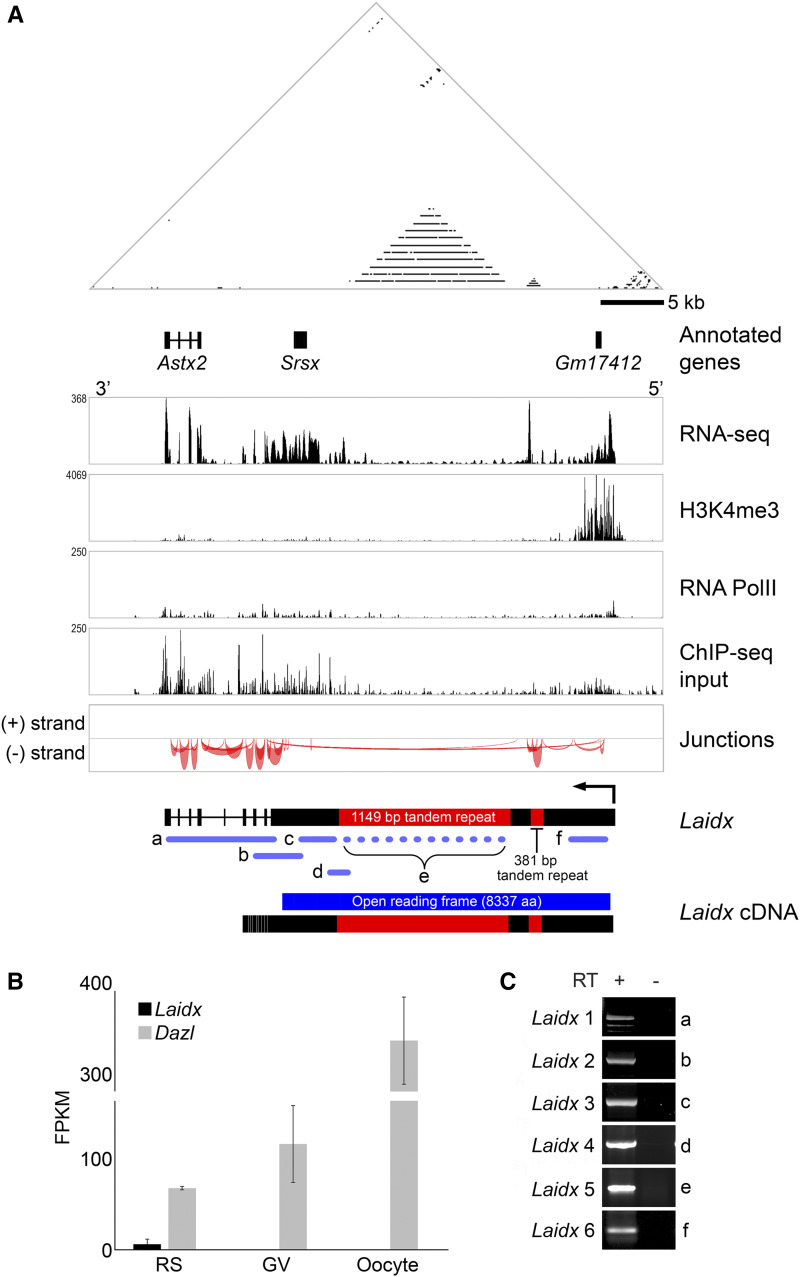
Figure 2.
A large transcription unit, Laidx, encompasses three partially annotated genes, including Srsx. (A) Self-symmetry triangular dot plot of a 45.5 kb region encompassing the large transcription unit. The coordinates of the chromosomal regions shown are chrX:123,443,060-123,488,539 (mm10). Positions of three partially annotated genes (Astx2, Srsx, and Gm17412) are indicated. Below the dotplot and gene annotations are aligned reads from RNA-seq performed on round spermatids, showing transcription extending from upstream of Gm17412 to the end of Astx2. In addition, ChIP-seq on round spermatids revealed modest enrichment of RNA polymerase II along the transcription unit and a small amount of enrichment at the transcription start site, along with broad enrichment of H3K4me3, a modification associated with active promoters. RNA-seq and ChIP-seq alignments were performed on repeat masked sequence (Smit et al. 2015). “Junctions” illustrates predicted splice sites based on RNA-seq. The height and thickness of the arcs are proportional to read depth spanning the junction, up to 50 reads. The predicted genomic organization of the Laidx gene is illustrated below. Purple bars represent select RT-PCR assays used to verify expression and are lettered to correspond with labels in (C). (B) Quantitation of RNA-seq data from round spermatids (RS), germinal vesicles (GV), and oocytes demonstrating transcription in the male but not female germline. Dazl, a gene expressed in both male and female germlines, is used as a control. FPKM, Fragments Per Kilobase per Million reads. (C) RT-PCR on RT (+) and no RT controls (-) performed on RNA isolated from adult testis.