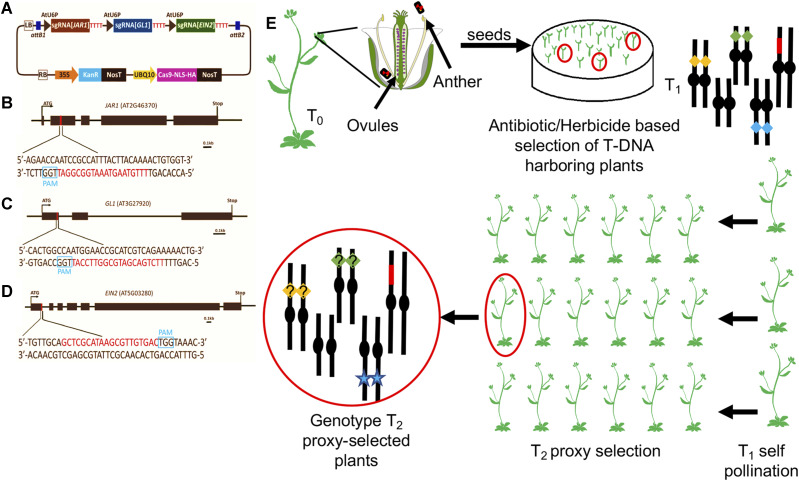
Figure 1.
Proxy-based selection scheme of CRISPR-mutagenized plants. (A) The pCUT3-CE construct contains three individual transcriptional units that generate sgRNA for JAR1, GL1 and EIN2 editing. The expression of each sgRNA is controlled by individual AtU6 promoters (U6P) and poly-T terminator (TTTTTT). (B-D) JAR1, GL1 and EIN2 target sites. Sequence in red denote the 20nt crRNA target site within each proxy gene. The PAM site is boxed in blue. Scale bar = 0.1kb. (E) Selection scheme using proxy plants: T0 plants are transformed with the binary vector shown in (A). In the T1 generation, each gene targeted is depicted as a yellow, green or blue diamond, each one positioned on a different Arabidopsis chromosome. The T-DNA harboring CRISPR-Cas9 is depicted as a red chromosome fragment. The genotype of proxy-selected T2 plants is shown at the left end of the scheme: the blue star depicts loss-of-function mutation of the proxy gene that allowed for the visual selection of edited plants, while yellow and green diamonds with a question mark depict surrogate genes of unknow allelic condition that need to be PCR-genotyped.