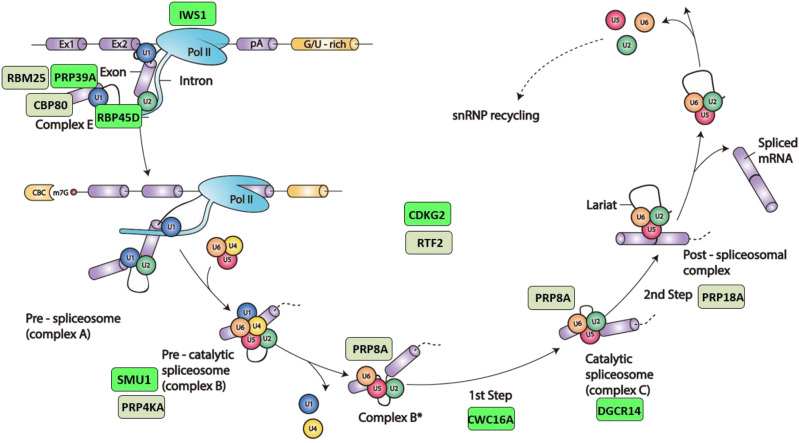
Figure 2.
Spliceosomal cycle and factors identified in the forward genetic screen. Splicing is catalyzed by the spliceosome, a large and dynamic ribonucleoprotein (RNP) machine located in the nucleus. Spliceosomes comprise five small nuclear (sn) RNPs, each containing a heptameric ring of Sm or Like-Sm proteins and a different snRNA (U1, U2, U4, U5 or U6), as well as numerous other non-snRNP proteins. During the splicesomal reaction cycle, the five snRNPs act sequentially on the pre-mRNA with a changing assemblage of non-snRNP proteins to form a series of complexes that catalyze two consecutive trans-esterification reactions. In complex E, U1 and U2 snRNPs first recognize the 5′ and 3′ splice sites branch points of introns and interact to form pre-spliceosomal complex A. The subsequent addition of preformed U4/U5/U6 tri-snRNP creates pre-catalytic complex B. Ensuing reorganization steps induce release of U1 and U4 snRNPs and conversion of complex B to complex B*, which catalyzes the first reaction yielding the free 5′ exon and lariat 3′-exon intermediates. The newly formed C complex catalyzes the second reaction to achieve intron lariat excision and exon ligation. Lastly, dismantling of the spliceosome frees individual components to assemble anew at the next intron. The positions of factors identified in our screen, as predicted by their orthologs in yeast or metazoans, are indicated by colored rectangles (green, Hyper-GFP; dull green, GFP-weak). Adapted by permission from Springer Nature, Nat. Rev. Mol. Cell Biol. 15: 108–121, A day in the life of the spliceosome, A. G. Matera and Z. Wang, 2014.