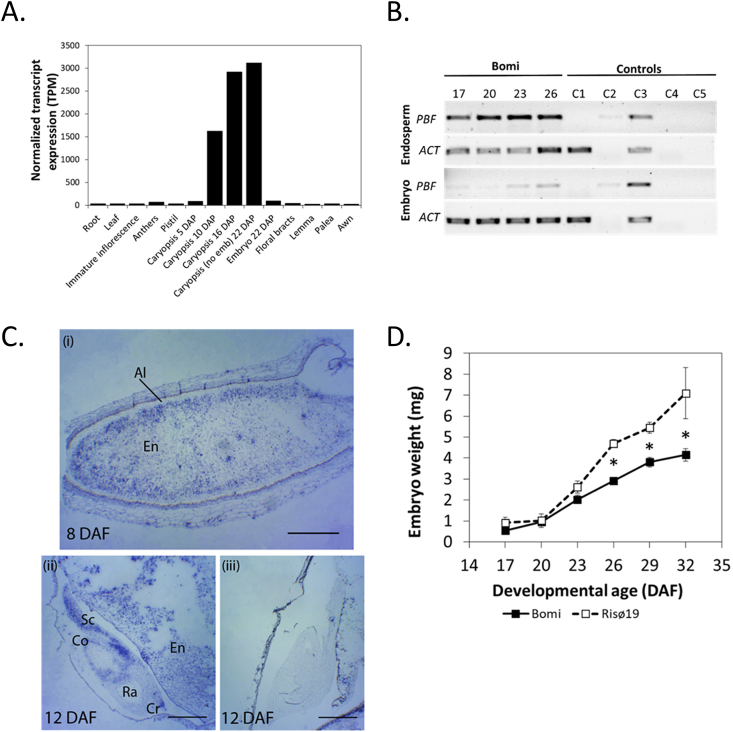
Fig. 4.
Patterns of expression of PBF/LYS3 in barley.
A. Tissue-specific expression according to the Barley eFP Browser 2.0 (www.bar.utoronto). Samples are from barley cv. Morex. Caryopsis (no Emb) means caryopsis without embryo. B. Temporal and tissue-specific patterns of LYS3/PBF expression in developing grains of wild type (Bomi) assessed by RT–PCR. Each cDNA sample was from a pool of 10–75 tissue samples each from an individual grain. Samples were collected from at least five spikes, each from a different plant. Amplicons were visualized on 1% agarose gels stained with SYBR Safe (Invitrogen, UK). Numbers above panels are DAF. PBF = Prolamin Binding Factor (HORVU5Hr1G048700). ACT = ACTIN.(HORVU1Hr1G047440), a constitutively expressed gene. Control reactions varied from the test reactions as follows. C1: contained cDNA from the barley PBF deletion mutant Risø18 at 26 DAF. No product was observed showing that the primers used were specific for PBF. C2: no DNase, no reverse transcriptase (RT). C3: no DNase. C4: no RT. C5: contained water used for PCR instead of cDNA. C. In situ localization of PBF in developing wild type (Bomi) grains at 8 and 12 DAF. (i) Longitudinal section of 8 DAF grain stained with antisense probe. PBF is expressed mostly in the starchy endosperm (En) cells. (ii) Longitudinal section of 12 DAF grain (including the embryo) stained with an antisense probe. PBF is expressed in the scutellum (Sc), coleoptile (Co) and coleorhiza tip (Cr). PBF is not expressed in the radicle (Ra). (iii) Longitudinal section of 12 DAF as in (ii) but stained with a sense probe (negative control). Scale bars are 0.5 mm (i) and 20 mm (ii and iii). D. Embryo fresh weight in developing grains of wild type (Bomi) and lys3 mutant Risø19. Embryos were extracted from developing grains and immediately weighed. Values are means ± SE (n > 4) for 10 embryos extracted from the middle of at least three spikes. Values are significantly different (p < 0.05, Tukey's HSD after one-way Anova, mutant compared to wild type) for 26, 29 and 32 DAF (as indicated by asterisk). Error bars are included for all data and if not visible are smaller than the marker.