Figure 2.
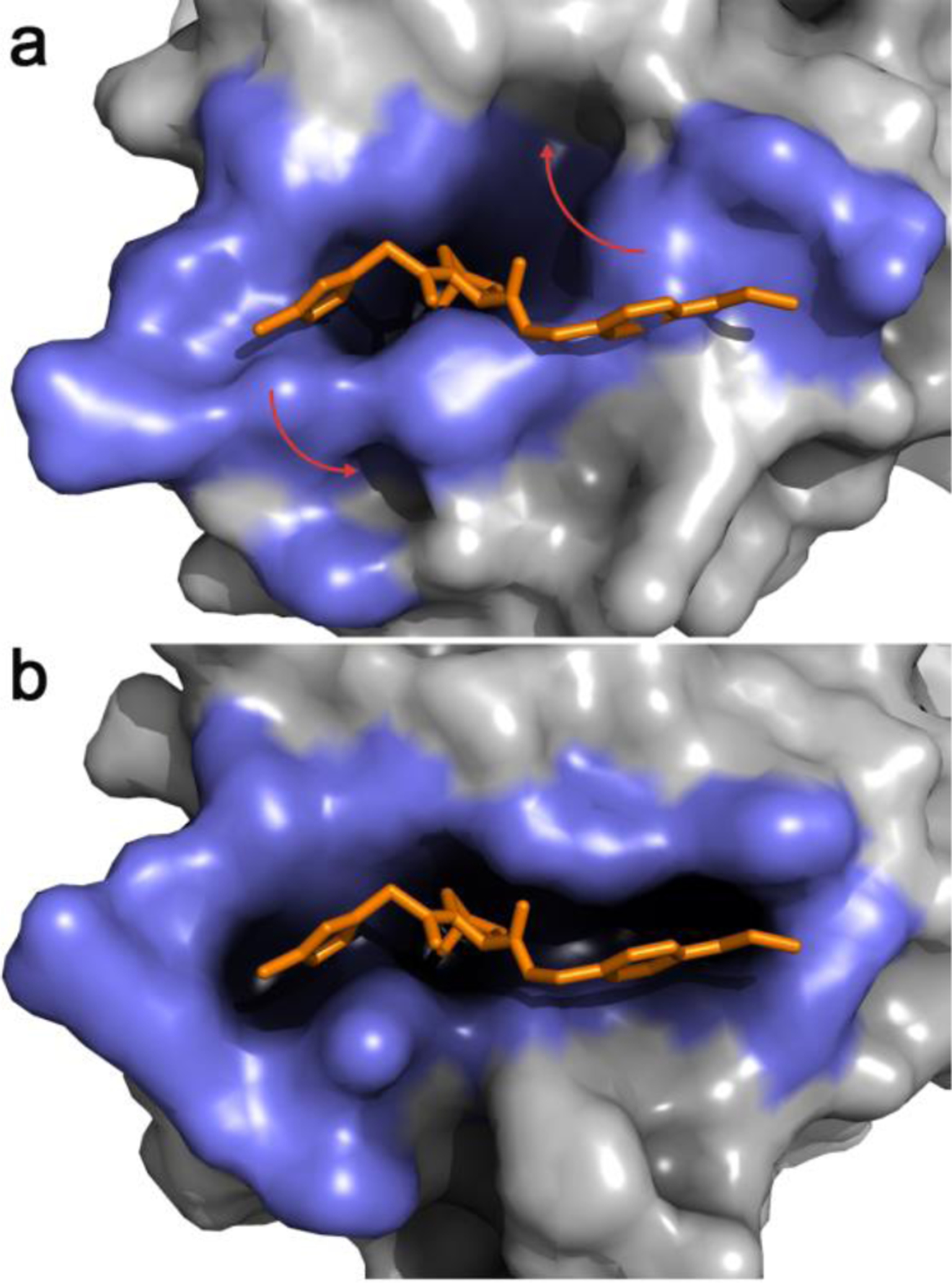
a. Representative snapshot from the most populated cluster of an equilibrium MD simulation of apo VHL. The binding-site lining residues (highlighted in slate blue) adopt a collapsed conformation with low druggability score b. A JEDI-biased MD simulation of apo VHL has rearranged the binding-site lining residues in a druggable conformation. In both panels an overlay of the crystallographic structure of a VHL ligand is displayed in orange sticks. Red arrows highlight the main structural changes.
