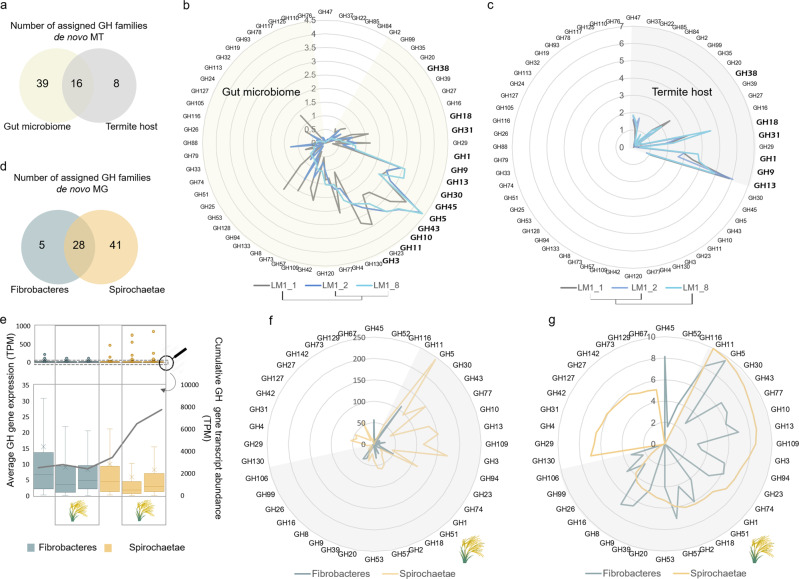
Fig. 3. Carbohydrate active enzymes (CAZymes) reconstructed from metatranscriptomic (MT) and metagenomic (MG) reads for the termite gut system.
a Venn diagram showing the number of assigned glycoside hydrolase (GH) families for the termite gut microbiome and host gut epithelium. b, c Comparison of the gene expression profiles (de novo MT, log2 transformed) for gene transcripts assigned to the different GH families and at the different stages of the Miscanthus feeding experiment, for the gut microbiome (b) and the host gut epithelium (c). d Venn diagram showing the number of assigned GH families for Fibrobacteres and Spirochaetae, based on the de novo MG reconstruction. e Average CAZyme genes expression and cumulative gene expression at the different stages of the feeding experiment and analysed separately for Fibrobacteres and Spirochaetae. Lower panel is a zoom on the gene expression profiles with the outliers (highly expressed genes; in some cases representing only partially reconstructed genes) removed. Boxes represent the interquartile range and error bars show the 95% confidence intervals (n = number of transcripts annotated as glycoside hydrolases). f, g Number of genes (f) and cumulative abundance of the most abundant GH families (g RNA-seq log2 transformed) at the time point LM1_8 (the end of the Miscanthus feeding experiment), and visualised separately for Fibrobacteres and Spirochaetae. Transcripts abundance (g) is calculated based on the RNA mappings (RNA-seq) to the MG contigs. Shaded parts correspond to shared GH families.