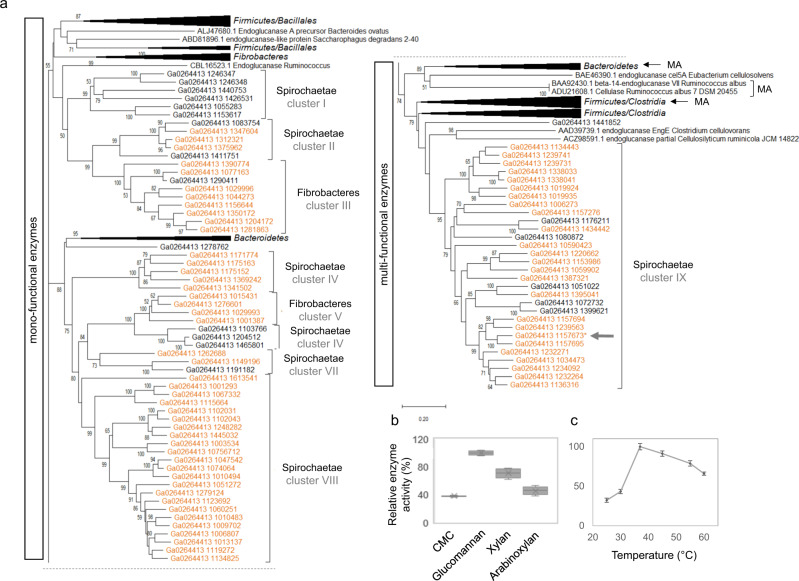
Fig. 5. Characterisation of the GH5_4 family.
a Unrooted neighbour-joining tree containing the de novo reconstructed genes from MG study (genes expressed under Miscanthus diet are highlighted in orange on the tree). Tree was cut in two parts along the dashed line. All GH5_4 characterised proteins were retrieved from the CAZY database and included on the tree. Clusters indicated with an arrow and designated as “MA” contain known multi-functional enzymes. The percentage of replicate trees in which the associated sequences clustered together in the bootstrap test (500 replicates) are shown next to the branches. Final alignment involved 157 amino acid sequences. Protein from the Spirochaetes cluster IX indicated with a grey arrow was heterologously produced and characterised. b Activity profiles for the heterologously produced and purified protein tested against CMC, glucomannan (galactomannan was negative, not displayed on the graph), xylan and arabinoxylan. Boxes represent the interquartile range and error bars show the 95% confidence intervals (n = 4). c Optimal temperature was assessed for glucomannan substrate. Error bars represent the standard deviation of a dataset (n = 3).