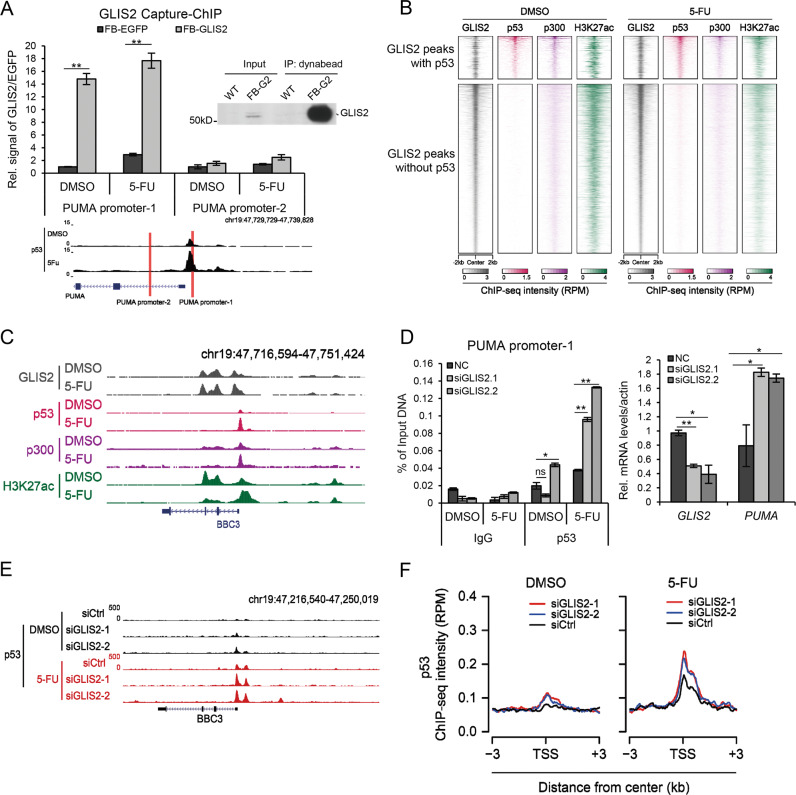
Fig. 3. GLIS2 represses p53 binding on PUMA promoter.
a CAPTURE-ChIP shows GLIS2 binding on PUMA relative to EGFP. The primer sets for qPCR are indicated at the bottom. b Heat maps generated from ChIP-seq data showing the occupancy of GLIS2, p53, H3K27ac, and p300 in HCT116. All rows are centered on the GLIS2 peaks. GLIS2 ChIP-seq was performed by Dynabeads MyOne streptavidin C1 (Thermo-Fisher 65001) in FB-GLIS2-expressing HCT116 stable cells. c The genome browser view of FB-GLIS2 on PUMA. d Cells were prepared as in 2A and ChIP analysis was performed with anti-p53 antibody. e The UCSC browser view shows p53 enrichment around PUMA after GLIS2 knockdown. f The average signals of p53 enrichment on p53 target genes (n = 523). *p-value ≤ 0.05, **p-value ≤ 0.01 (t-test). Histograms are presented as mean ± s.d. of three biological replicates.