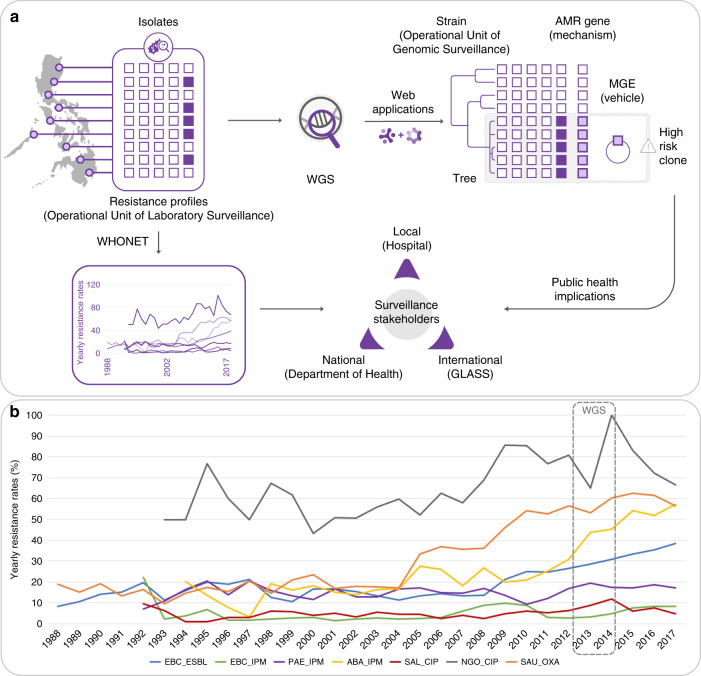
Fig. 1. Implementing whole-genome sequencing (WGS) for AMR surveillance in the Philippines.
a ARSP workflow and enhanced detection of high-risk clones by WGS. Isolates collected by sentinel sites are tested for susceptibility to antibiotics (open squares: susceptible, solid squares: resistant). The data are stored as resistance profiles in WHONET and summaries of resistance trends are shared yearly with surveillance stakeholders. WGS of bacterial isolates and interpretation with web applications like Microreact and Pathogenwatch provide information on genetic relatedness (strains), known AMR determinants (mechanisms), and the mobile genetic elements (MGE, vehicles) for their dissemination, thus allowing us to detect high-risk clones. b Detail of trends in antimicrobial resistance in the Philippines. Yearly resistance rates for key bug–drug combinations based on phenotypic data collected by sentinel sites. EBC Enterobacteriaceae (K. pneumoniae, E. coli, Salmonella enterica), PAE P. aeruginosa, ABA A. baumannii (Acinetobacter spp. before the year 2000), SAL Salmonella enterica, SAU S. aureus, ESBL extended-spectrum beta-lactamase production suspected, or non-susceptible to the following antibiotics IPM imipenem, CIP ciprofloxacin, OXA oxacillin, WGS box: period covered by the retrospective sequencing survey.