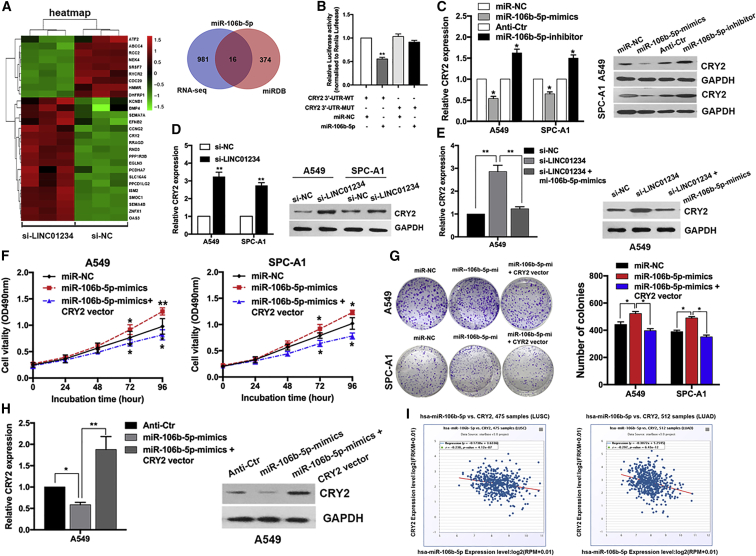
Figure 6.
CRY2 Is a Key Target of miR-106b-5p in NSCLC Cells
(A) Hierarchically clustered heatmap of upregulated and downregulated genes in LINC01234-depleted A549 cells. Venn diagram shows the overlap between predicted miR-106b-5p targets by the miRDB database and LINC01234 depletion upregulated genes. (B) Luciferase activities were measured in NSCLC cells cotransfected with the luciferase reporter containing CRY2 wild-type or mutant and the mimics of miR-106b-5p. (C and D) qPCR and western blot analysis of CRY2 mRNA and protein expression after transfection with miR-106b-5p mimics or inhibitors (C) and LINC01234 siRNA (D). (E) qPCR and western blot analysis of CRY2 mRNA and protein expression after cotransfection with LINC01234 siRNA and miR-106b-5p mimics. (F) Growth curves show the proliferation ability of miR-106b-5p mimics and the CRY2 overexpression vector cotransfected cell. (G) Colony formation assays were used to evaluate the colony formation capacity of miR-106b-5p mimics and the CRY2 overexpression vector cotransfected cell. (H) qPCR and western blot analysis of CRY2 mRNA and protein expression after cotransfection with miR-106b-5p mimics and the CRY2 overexpression vector. (I) The scatterplot images show the correlation between miR-106b-5p and CRY2 in TCGA dataset.∗p < 0.05, ∗∗p < 0.01.