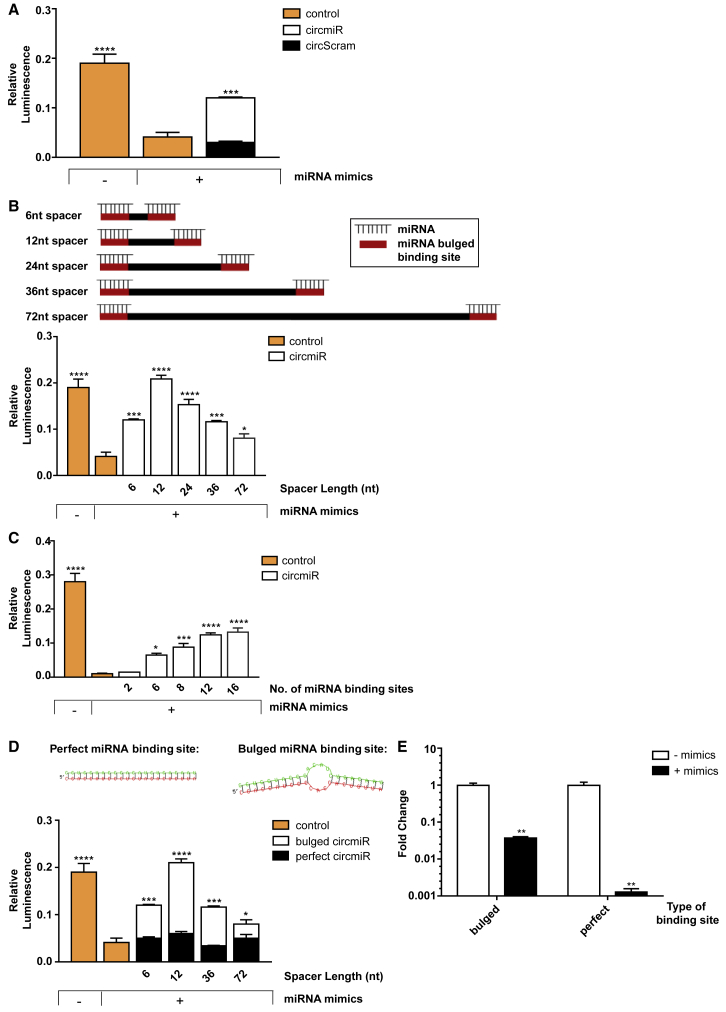
Figure 2.
Engineered circmiRs Are Efficient Sponges of miR-132 and -212
Luciferase rescue reporter assays using dual reporter constructs with miR-132 and -212 binding sites inserted into the 3′ UTR of the Renilla luciferase gene. HEK293T cells were co-transfected with dual reporter plasmid psiCheck2, miR-132 and -212 mimics, and respective circRNA expression constructs for 48 h, to determine (A) the effect of circmiR versus circScram, (B) the effect of circmiR with different spacer lengths as follows: 6, 12, 24, 36, and 72 nucleotides, (C) the effect of circmiR with different numbers of miRNA binding sites as follows: 2, 6, 8, 12, 16, and (D) the effect of bulged versus perfect complementary miRNA binding sites. RNAhybrid prediction of structures are shown for each binding site type upon miRNA binding. (n = 3); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 relative to control with mimics. One-way ANOVA with Benjamini-Hochberg adjustment. (E) Expression abundance 48 h post-transfection of circmiRs carrying either 12 bulged or perfect miRNA binding sites in the presence or absence of mimics in HEK293T cells using qPCR. (n = 3); ∗∗p < 0.01 relative to control without mimics. Student’s t test.