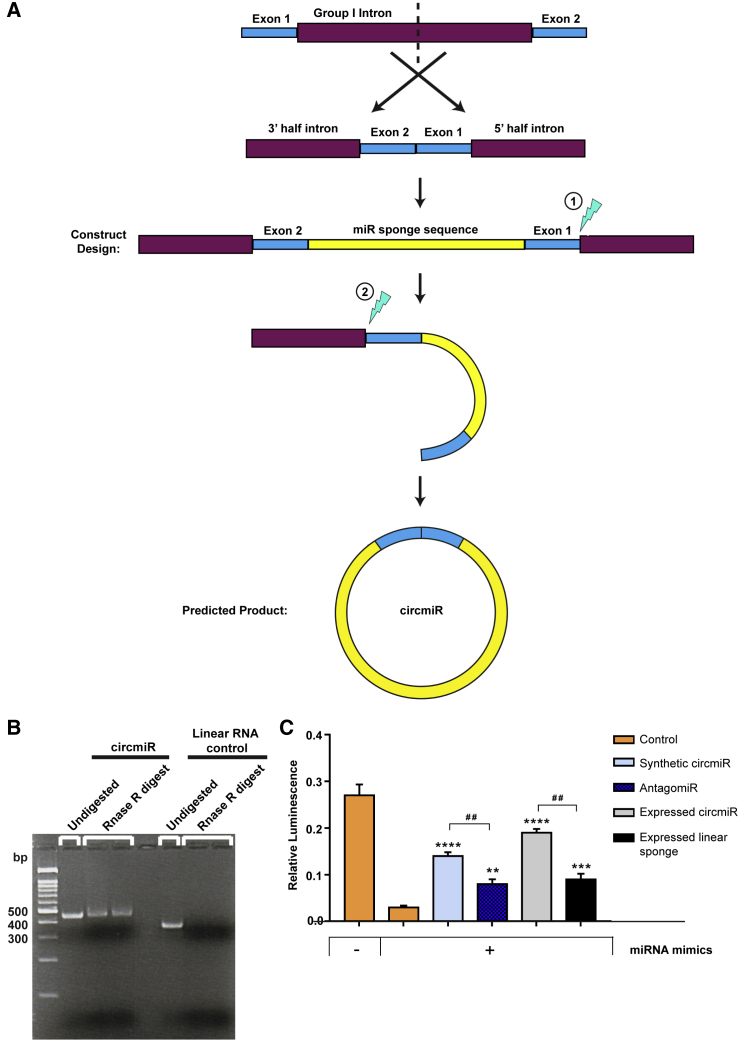
Figure 4.
Engineering of Synthetic circmiRs In Vitro Using Group I Permutated Intron-Exon Splicing
(A) Schematic illustration of permutated intron-exon synthetic circmiR construct design and predicted splice product. Partial sections of the group I intron of T4 bacteriophage Td gene are reversed to allow the intron to fold into a stabilized structure required for splicing. Downstream exon 2 fragment is ligated to upstream exon 1 fragment. The miRNA sponge sequence is inserted between the exon 2-exon 1 junction. During autocatalytic splicing, a two-step transesterification reaction (lightning symbol) occurs that results in ligation of both ends of the miRNA sponge sequence. (B) RNase R digests separated on agarose gel to confirm circmiR circularization. (C) Luciferase reporter assays using dual reporter constructs with miR-132 and -212 binding sites inserted into the 3′ UTR of Renilla. HEK293T cells were co-transfected with dual reporter plasmid psiCheck2, miR-132 and -212 mimics, and respective constructs to determine the effect of antagomiRs versus circmiRs. (n = 3); ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 relative to control with mimics. One-way ANOVA with Benjamini-Hochberg adjustment. ##p < 0.01 as indicated. Student’s t test.