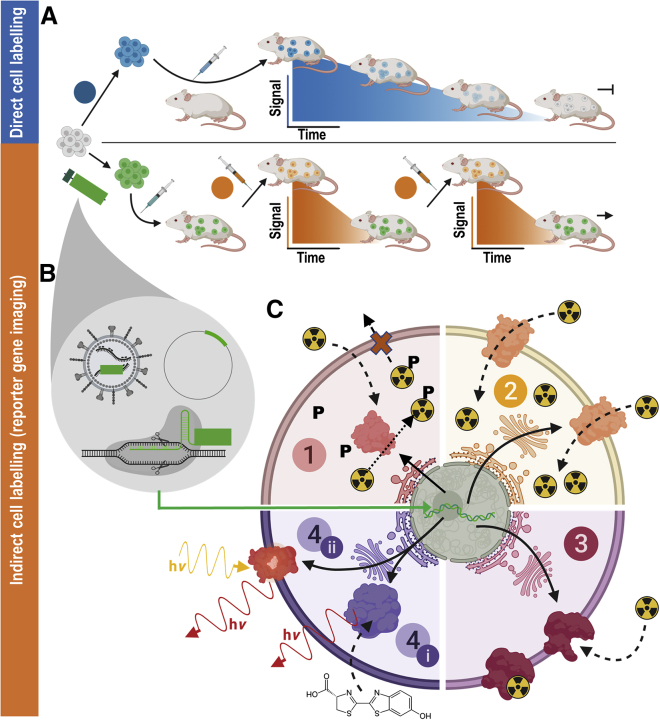
Figure 1.
In Vivo Cell Tracking Using Reporter Genes
(A) (Blue) Direct cell labeling employs ex vivo-labeled cells that are administered to animals and can be tracked until cells lose their labels (depicted using blue signal versus time cartoon plots), e.g., through label efflux, via label dilution in fast-growing cells, or radioisotope decay if radiotracers are used. (B) (Orange) Indirect cell labeling requires cells that have been genetically manipulated to express a reporter gene (green). The genetic engineering options frequently employed in reporter gene applications include viruses (e.g., lentiviruses, γ-retroviruses), gene editing, or episomal plasmids (see cartoons within gray drop). The cells are imaged using the features of the reporter gene, which renders the cells traceable in vivo. Cells are detected in vivo through molecular probe administration (depicted using orange signal versus time cartoon plots); if radiotracers are used, their half-life is short to enable short repeat-imaging intervals and keep administered doses low. Reporter gene imaging does not suffer from label dilution in fast-growing cells and hence permits much longer, theoretically indefinite observation times. (C) Molecular imaging mechanisms of frequently used reporter genes. (1) Enzymes entrapping molecular probes (light red): these reporter enzymes entrap a substrate that is already detectable by imaging. A frequent mechanism for this entrapment relies on phosphorylation of a substrate that has either actively or passively entered the cell, and upon phosphorylation can no longer leave the cell. Examples are nucleoside kinases such as HSV1-tk. (2) Transporter proteins (yellow): these reporters are expressed at the plasma membrane of cells, and each expressed reporter can transport several labeling agent molecules into the cell, which constitutes a signal amplification mechanism. The radionuclide transporters NIS and NET belong to this class of reporters. (3) Cell surface molecules (pink): these reporters are expressed at the plasma membrane of cells, and molecular probes bind directly to them; minor levels of signal amplification are theoretically possible if several labels bind directly to each reporter protein, or if several labels could be fused to a reporter binding molecule; however, signal amplification is inferior compared to transporters, and often they are used with a 1:1 stoichiometry. Examples for this reporter class are tPSMAN9Del and SSTR2. (4) Signal generating proteins (purple). (i) Enzyme-based reporters bind to their substrate and catalyze the production of a detectable signal. Examples are luciferases, which convert an externally supplied chemical substrate into detectable light (hν). (ii) Fluorescent proteins contain an intrinsic fluorescence-generating moiety if appropriately excited by light. Fluorophore excitation results in emission of detectable longer wavelength/red-shifted light. For details and literature references to relevant reporter genes, see Tables 1 and 2. The figure was generated using Biorender.com.