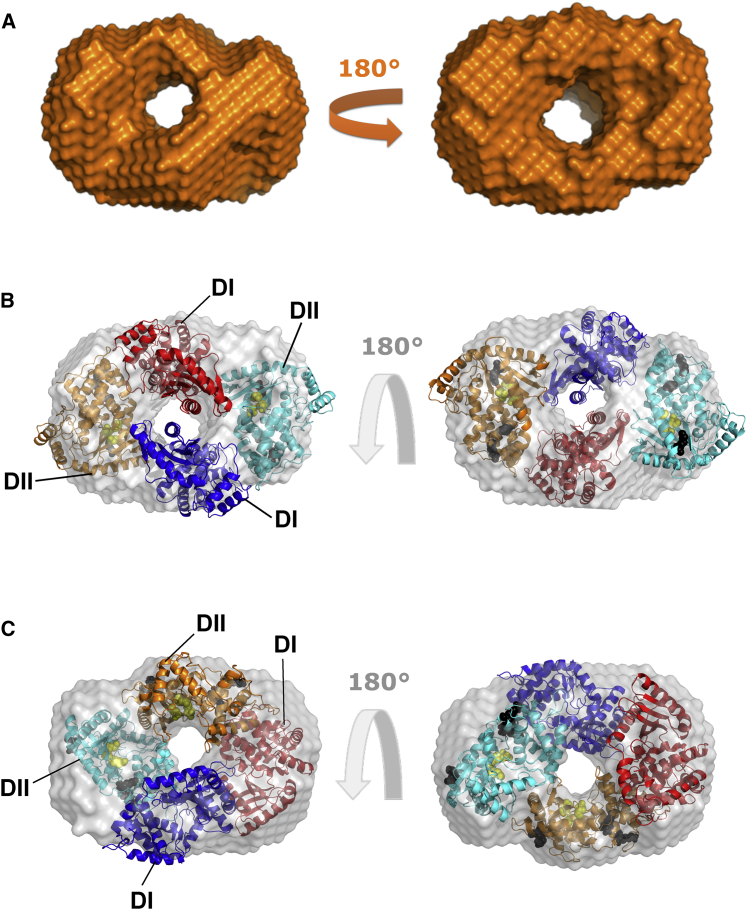
Figure 3.
OAS2 ab initio modeling. (A) The DAMMIN ab initio model for OAS2 is given, showing protein surface representation and rotation at 180°. (B) Superimposition of the atomistic structure of OAS2 calculated using the program CORAL on the averaged-filtered ab initio low-resolution structure obtained from DAMMIN and DAMFILT is shown. (C) Superimposition of the atomistic structure of OAS2 calculated using the program CLUSPRO on the averaged-filtered ab initio low-resolution structure obtained from DAMMIN and DAMFILT is shown. In (B) and (C), the dsRNA binding sites are represented by black spheres, whereas the aspartic acid residues are indicated by yellow spheres. To see this figure in color, go online.