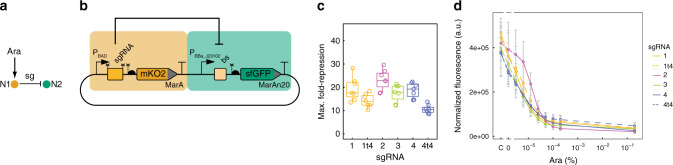
Fig. 1. Assessment of CRISPRi repression through a 2-node NOT logic circuit.
a Schematic representation of the circuit architecture, where the first node (N1) is induced by Ara and in turn produces a sgRNA (sg) that represses the second node (N2). b Details of the circuit design. In addition, AraC, dCas9, and Csy4 are constitutively expressed. Bent arrows: promoters; squares: sgRNA binding sites; rectangles: sgRNAs; crosses: csy4 recognition sites; semicircles: RBSs; pointed rectangles: reporter genes with degradation tags; and T-s: transcriptional terminators. c Maximum fold-repression of N2 achieved by four different sgRNAs, calculated as the GFP fluorescence of a fully-induced (0.2% Ara) circuit relative to a control lacking the sgRNA. Truncated variants of sgRNA-1 and -4 lacking the four 5′ nucleotides (t4) were also assessed. Data correspond to seven biological replicates. Boxes represent quartiles Q1 (box lower limit), Q2 (i.e., median, internal line) and Q3 (box upper limit); whiskers comprise data points that are no more than 1.5× IQR (inter-quartile region, i.e., the length of the box) away from the box. d Dose-response curves for the NOT circuits operating through sgRNA-1 to -4 (including t4 variants of sgRNA-1 and -4). The response (GFP fluorescence) of all four circuits can be modulated by adjusting the input dose (Ara). The controls lacking the sgRNAs (C) showcase some level of PBAD leakiness in the NOT circuits that results in weak repression even in the absence of Ara (0). Mean and s.d. of three biological replicates. Source data are provided as a Source Data file.