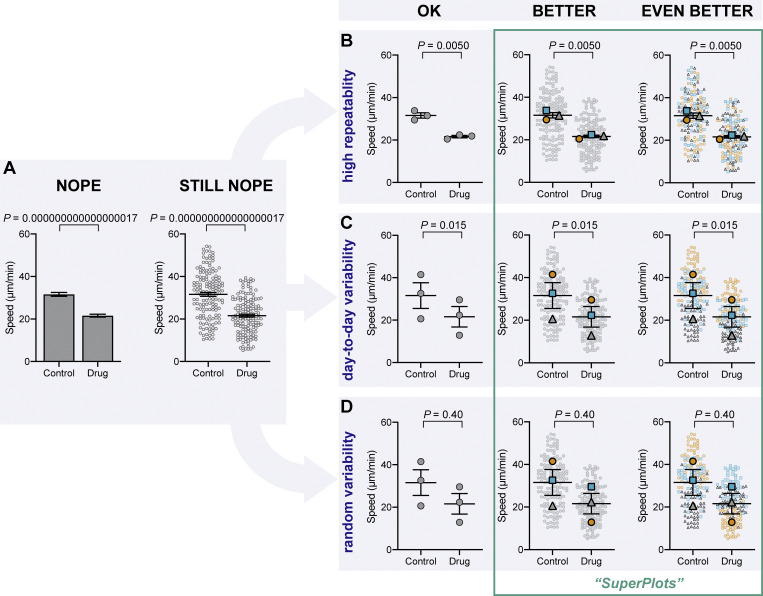
Figure 1.
The importance of displaying reproducibility. Drastically different experimental outcomes can result in the same plots and statistics unless experiment-to-experiment variability is considered. (A) Problematic plots treat n as the number of cells, resulting in tiny error bars and P values. These plots also conceal any systematic run-to-run error, mixing it with cell-to-cell variability. (B–D) To illustrate this, we simulated three different scenarios that all have identical underlying cell-level values but are clustered differently by experiment: B shows highly repeatable, unclustered data, C shows day-to-day variability, but a consistent trend in each experiment, and D is dominated by one random run. Note that the plots in A that treat each cell as its own n fail to distinguish the three scenarios, claiming a significant difference after drug treatment, even when the experiments are not actually repeatable. To correct that, “SuperPlots” superimpose summary statistics from biological replicates consisting of independent experiments on top of data from all cells, and P values were calculated using an n of three, not 300. In this case, the cell-level values were separately pooled for each biological replicate and the mean calculated for each pool; those three means were then used to calculate the average (horizontal bar), standard error of the mean (error bars), and P value. While the dot plots in the “OK” column ensure that the P values are calculated correctly, they still fail to convey the experiment-to-experiment differences. In the SuperPlots, each biological replicate is color-coded: the averages from one experimental run are yellow dots, another independent experiment is represented by gray triangles, and a third experiment is shown as blue squares. This helps convey whether the trend is observed within each experimental run, as well as for the dataset as a whole. The beeswarm SuperPlots in the rightmost column represent each cell with a dot that is color-coded according to the biological replicate it came from. The P values represent an unpaired two-tailed t test (A) and a paired two-tailed t test (B–D). For tutorials on making SuperPlots in Prism, R, Python, and Excel, see the supporting information.