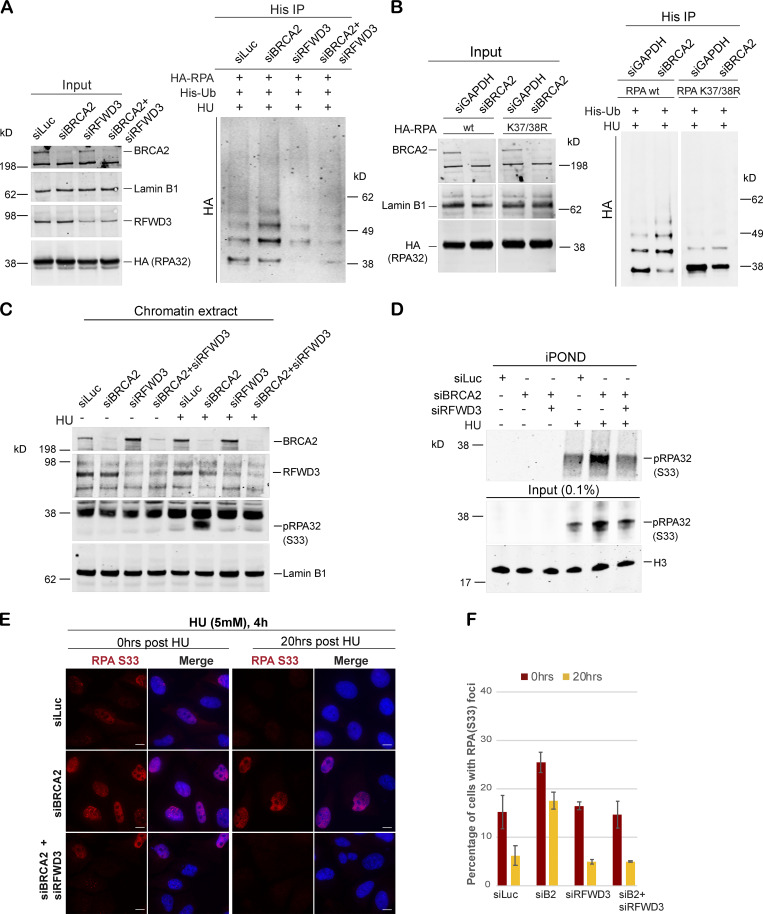
Figure 5.
Hyperubiquitination of RPA in BRCA2-deficient cells undergoing replication stress is performed by the E3 ligase RFWD3. (A) Immunoprecipitation analysis of RPA ubiquitination in HEK293T cells transfected with control siRNA (siLuc) or siRNA for BRCA2, RFWD3, or both BRCA2 and RFWD3. HEK293T cells with indicated siRNAs were transfected with His-tagged ubiquitin and HA-tagged RPA32. Cells were treated with 5mM HU and harvested 3 h after treatment. His immunoprecipitation was done as indicated above, and blots were immunoblotted with HA. (B) Lysine K37 and K38 amino acid residues in RPA get ubiquitinated by RFWD3 in BRCA2-depleted cells. Cells were transfected with indicated siRNAs, followed by transfection with HA-tagged WT or K37/38R RPA mutant. Cells were processed for His-immunoprecipitation as described above. (C) Western blot analysis of RPA32 accumulation on chromatin after disrupting RPA ubiquitination by codepletion of RFWD3 in BRCA2-depleted cells. U2OS cells transfected with indicated siRNAs were treated with 5 mM HU and harvested 3 h after treatment. Chromatin extracts were prepared, and relevant Western blot was probed with pRPA32 (S33). (D) Western blot analysis of cells with indicated siRNA for input and captured proteins isolated by iPOND. Cells were pulse labeled with EdU for 10 min followed by treatment with 5 mM HU and harvested 3 h after treatment. (E and F) IF analysis to study the effect of RPA ubiquitination on RPA32 accumulation in BRCA2-deficient cells. Scale bars in E indicate 10 µm. (E) U2OS cells transfected with indicated siRNAs were treated with 5 mM HU for 4 h and fixed right after (0 h) or 20 h after treatment. Cells were stained with pRPA32-S33. (F) Graph indicates percentage of cells with pRPA-S33 foci after HU induced DNA damage. Error bars indicate SD (n = 3).