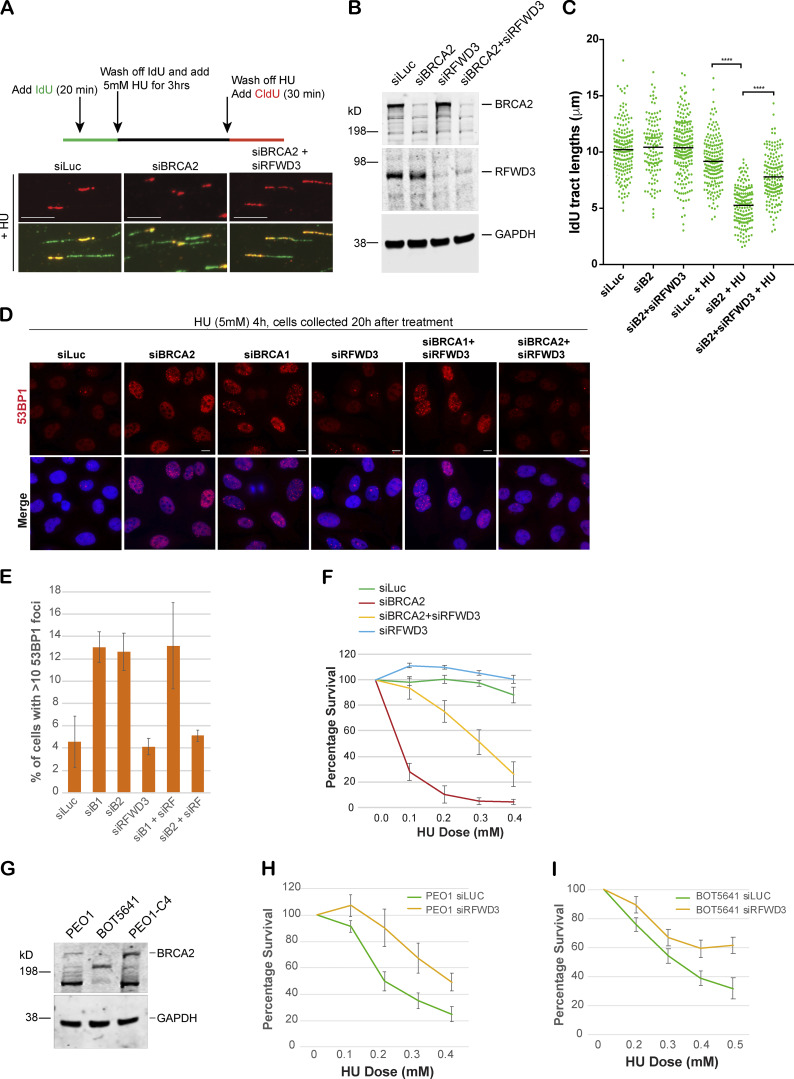
Figure 6.
RFWD3 depletion rescues fork degradation, fork collapse, and cell sensitivity to stalled fork–inducing agents in BRCA2-depleted cells and BRCA2 mutant tumor cells. (A) Top: Schematic of how DNA fiber experiment was performed. Bottom: Representative tracts from DNA fiber experiments with U2OS cells transfected with indicated siRNAs. Green and red tracts correspond to IdU and CldU incorporation, respectively. Scale bars indicate 10 µm. (B) Western blot analysis of whole-cell lysate to confirm knockdown of BRCA2 and RFWD3 after transfecting cells with indicated siRNAs. These extracts are from the cells used for Fiber assay. (C) Scatterplots compare the tract lengths of IdU-labeled fibers between different siRNA conditions and in the presence or absence of HU, with black lines indicating the median. ****, P < 0.0005. (D and E) IF and graph of 53BP1 recruitment in U2OS cells transfected with indicated siRNAs. Cells were treated with 5 mM HU for 4 h and then collected 20 h after treatment. Graph indicates the percentage of cells with more than 10 53BP1 foci per cell. Scale bars in D indicate 10 µm. (F) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of U2OS cells transfected with indicated siRNAs to HU. HU was added for 4 d, and cells were allowed to recover for 2 d before harvesting them for CellTiter-Glo–based analysis. (G) Western blot analysis of total BRCA2 protein levels in different BRCA2 tumor lines. PEO1 is an ovarian cancer cell line that has a BRCA2 homozygous mutation 5193C>G, which would normally result in a stop codon at amino acid 1655. BOT5641 is a BRCA2 breast tumor line that has a c.6486_6489del mutation. (H and I) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of BRCA2 mutant tumor lines PEO1 (H) and BOT5641 (I) to HU after depletion of RFWD3. Error bars indicate SD between triplicates.