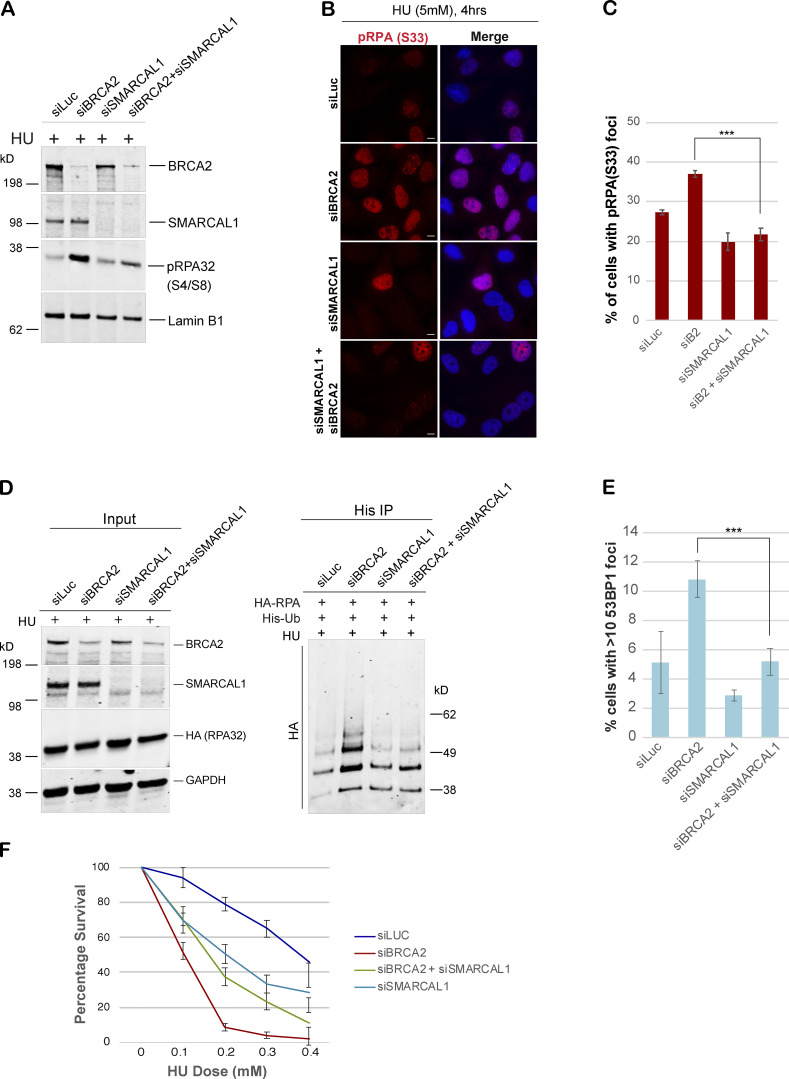
Figure 8.
SMARCAL1-mediated fork reversal is required for accumulation of hyperubiquitinated RPA in BRCA2-deficient cells. (A) Western blot-based analysis of pRPA32 accumulation after disrupting reversed fork formation by codepletion of SMARCAL1 in BRCA2 deficient cells. Experimental conditions used are as described above. Nuclear extracts were prepared. Blot was probed with anti-pRPA32 (S4/S8). (B and C) IF and graphs of pRPA32 (S33) recruitment in U2OS cells transfected with the indicated siRNAs. Cells were treated with 5 mM HU and harvested 4 h after damage. ***, P < 0.005. Statistical significance was determined by the two-tailed Student’s t test and the error bars indicate SD (n = 3). Scale bars in B indicate 10 µm. (D) Immunoprecipitation analysis of RPA ubiquitination in HEK293T cells transfected with indicated siRNAs. Experimental conditions used are as described above. (E) Quantification of IF-based analysis of 53BP1 foci in U2OS cells transfected with indicated siRNAs. Experimental conditions used are as described above. Graph indicates the percentage of cells with >10 53BP1 foci per cell. ***, P < 0.005. (F) CellTiter-Glo–based cell survival assay was used to determine the sensitivity of U2OS cells to HU in cells depleted of BRCA2, SMARCAL1, and/or BRCA2+SMARCAL1. Error bars indicate SD between triplicates. Experimental conditions used are as described above.