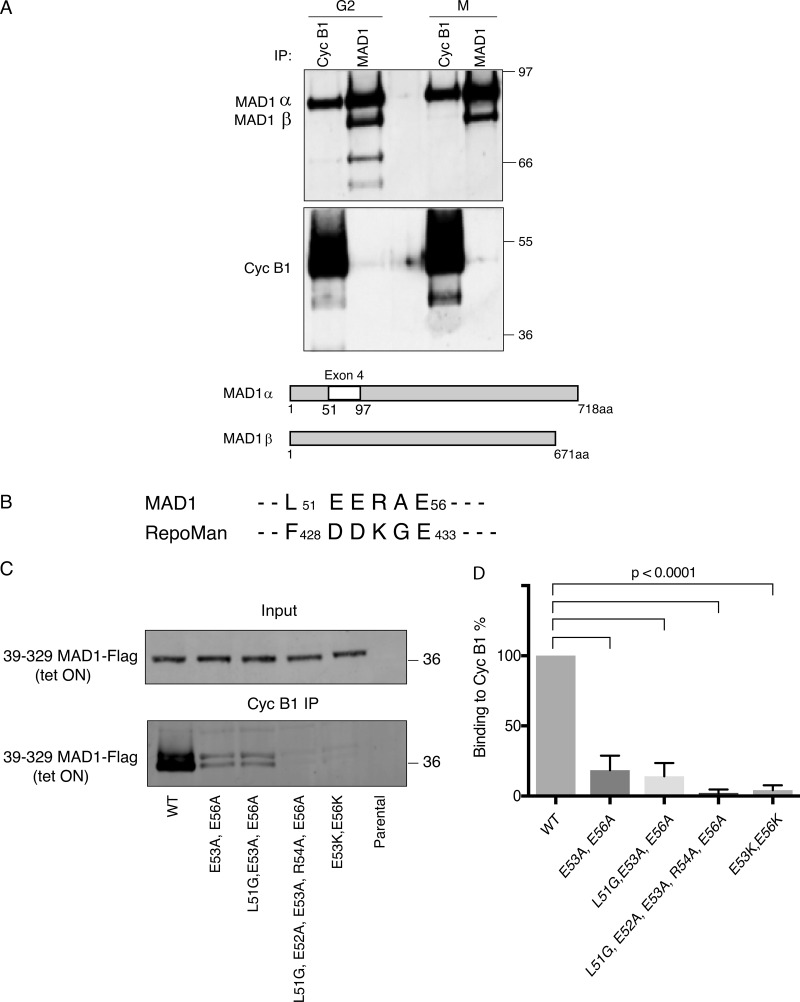
Figure 1.
MAD1 binds to cyclin B1 through the acidic face of a helix within exon 4. (A) HeLa cells were synchronized in either G2 phase or mitosis (M), cyclin B1 and MAD1 were immunoprecipitated (IP), subjected to SDS-PAGE and immunoblotted with anti-MAD1 (upper panel), and anti-cyclin B1 antibodies (lower panel). The difference in stoichiometries of binding is likely to be related to epitope accessibility, but we note that Schweizer et al. (2013) previously identified cyclin B1 in MAD1 immunoprecipitates. Schematic shows the location of exon 4 in MAD1α that is absent from MAD1β. (B) Similarity between MAD1 and Repoman sequences within the regions found to interact with cyclin B1. (C) HeLa cells expressing wild-type or mutated MAD1-Flag (39-329aa) from a tetracycline-inducible promoter (tet ON) were synchronized in either G2 phase or mitosis 12 h after adding tetracycline. Cyclin B1 was immunoprecipitated, subjected to SDS-PAGE, blotted with anti-FLAG antibody, and assayed on a LiCOR Odyssey scanner. (D) The data from three experimental repeats were normalized to the amount of cyclin B1 binding to wild-type MAD1 and plotted using Prism software. Error bars represent SD; two-tailed P values were calculated using an unpaired t test.