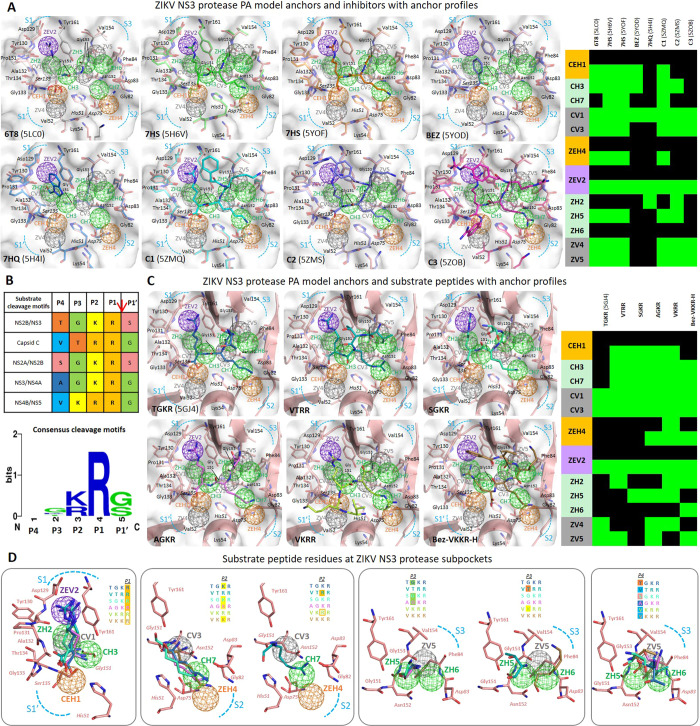
Figure 3.
ZIKV NS3 protease PA model anchors with inhibitors and substrate peptides. (A) The crystal structures of eight inhibitor-bound ZIKV NS3 proteases overlapped with the protease PA model depicting active site subpockets (surface), anchors (mesh spheres), residues and inhibitors (both shown in sticks). The anchor occupancy profiles of inhibitors are shown as heat-map (green – anchor interaction, black - no interaction). (B) Summary of ZIKV NS3 protease substrate cleavage motifs in the genome polyprotein showing P4-P3-P2-P1↓P1′ residues; Web logo showing consensus residues at cleavage motifs. (C) ZIKV NS3 protease PA model with P4-P1 substrate peptides, crystal structure for bound peptide TGKR (5GJ4) and docked poses for substrate peptides VTRR, SGKR, AGKR, VKRR and Bez-VKKR-H are displayed; the anchor profiles of the substrate peptides are displayed as heat-map. (D) At each subpocket, the binding substrate peptide residues P1, P2, P3 and P4 are seen occupying the anchors.