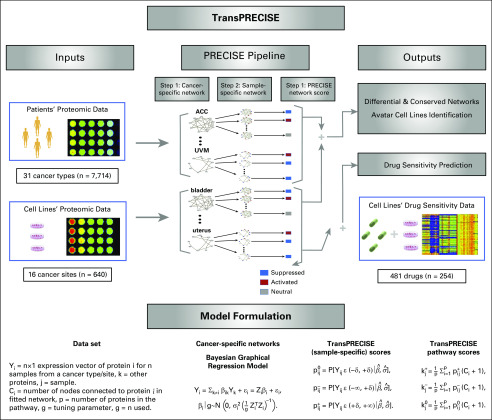
FIG 1.
Overview of the TransPRECISE framework. The first step of TransPRECISE involves implementing the PRECISE pipeline on two sets of RPPA protein expression data –namely, cancer patients (7,714 samples across 31 different cancer types) and cancer cell lines (640 samples across 16 different cancer tissues). For each combination of 47 cancer types across cell lines and patients and the 12 pathways, the PRECISE procedure is executed in three consecutive steps: fitting cancer-specific protein networks using Bayesian graphical regression (step 1); deconvolving these cancer-specific networks to fit sample-specific pathways networks (step 2); and aggregating the sample-specific networks to obtain calibrated TransPRECISE scores and pathway activity status (step 3). The cancer-specific networks from step 1 are compared across patients and cell lines for each pathway for pan-cancer identification of differential and conserved pathway activities. The TransPRECISE scores from steps 2 and 3 are used to identify potential avatar cell lines and the lineages for patient tumors and to construct prediction models for drug sensitivity trained in in vivo drug sensitivity and used for in silico drug sensitivity prediction of patients′ drug response. The bottom panel provides the details and equations for the computational steps of the Bayesian graphical regression procedure and post-processing of the regression outputs to obtain the cancer-specific and sample-specific summaries. All probabilities are computed under the fitted Bayesian graphical regression model with the estimated parameters, with the superscripts 0, +, and −, respectively corresponding to the neutral, activated, or suppressed status of the pathway. The pijs are the posterior probabilities corresponding to protein i and sample j, and the kjs are the aggregated pathway scores for sample j.