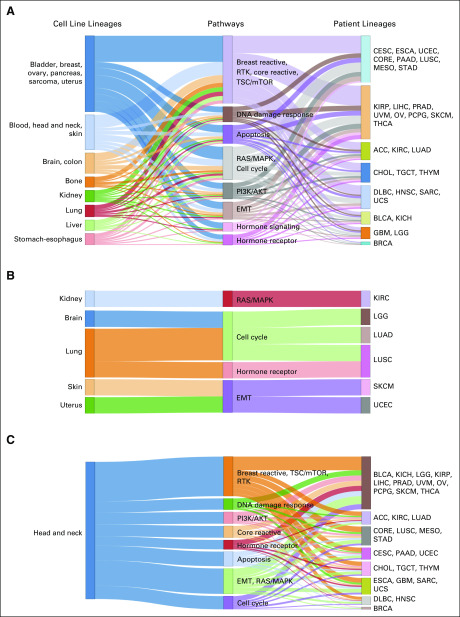
FIG 3.
Sankey diagrams for patient and cell line cancers with conserved pathway-specific connectivity. (A) The columns contain cell line cancers, pathways, and patient cancers from left to right, respectively. A cell line cancer tissue is connected to a pathway if the connectivity score (CS) for that cancer type-pathway pair (defined as the proportion of edges out of all possible undirected edges in the pathway that are held by that cancer type) is more than 900 out of 1,000 random CS values computed for that cancer type, with repeated random selection of the same number of proteins as in the pathway from the pool of all proteins across the 12 pathways. The connection between a patient cancer type to a pathway is also determined by the same rule. The length of the middle (pathway) column pieces indicate the participation of that pathway in driving the conservation across the two model systems. As seen in panel A, ovary and uterus cell lines were connected via the hormone signaling (breast) pathway with BRCA; lung, kidney, and stomach-esophagus cell lines were linked together with two clusters of patient cancers (KICH, KIRP, PRAD, LGG and LUSC, UCEC, STAD) via the RTK pathway. (B) The Sankey diagram contains only the subset of cell line cancer (ie, patient cancer pairs that have same tissue-specific lineage), and the cutoff for CS values is higher than 800 of the 1,000 random CSs obtained using the random selection of proteins. Panel B presents clear confirmations of conservation of activities across model systems within cancer tissues, some specific examples being bladder-core reactive (BLCA), kidney (RTK-KICH and KIRP), kidney-hormone receptor (KIRC), ovary-hormone signaling (OV), and stomach-hormone receptor (ESCA and STAD). (C) The Sankey diagram contains only the subset of the edges that are originating from the head and neck cancer cell line type, and the cutoff for CS values is higher than 800 of the 1,000 random CSs obtained using the random selection of proteins.