FIGURE 8.
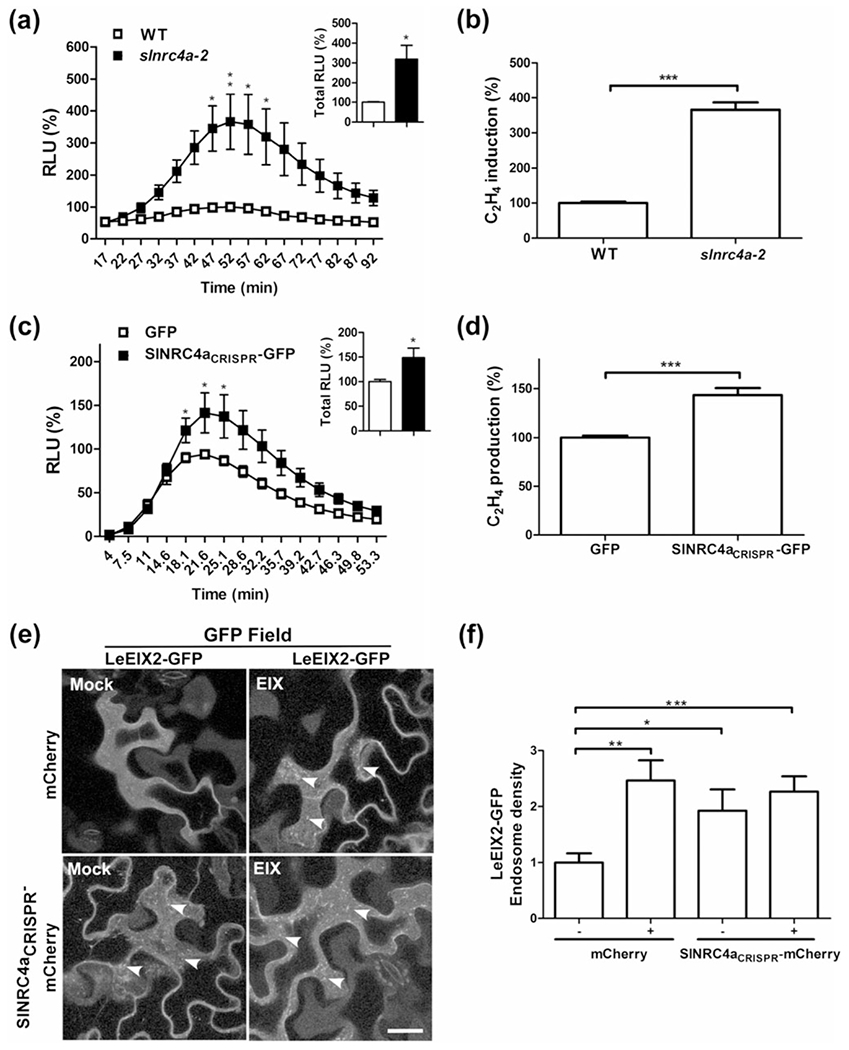
Effect of SlNRC4a CRISPR-Cas9 tomato editing on defence responses. (a) Leaf disks of slnrc4a-2 CRISPR-Cas9 edited or WT plants (M82 control) were taken from leaves 5-7. Leaf disks were floated on water for ~4 hr and then replaced with luminescence solution containing 1 μg ml−1 of EIX. Luminescence (RLU) was immediately measured to track the ROS burst. Average ± SEM values of three independent experiments, n = 12 each. Asterisks indicate significant differences with control treatment (two-way analysis of variance, **P < 0.001, *P < 0.05). (b) Leaf disks of slnrc4a-2 CRISPR-Cas9 edited or WT tomato plants (M82 control) were taken from leaves 5-7. Leaf disks were floated on a 250 mM of sorbitol solution with 1 μg mL−1 of EIX. Ethylene biosynthesis was measured after 5 hr. Ethylene induction was defined as the ΔEthylene (Ethylene+EIX – Ethylene−EIX). Control induction average value was defined as 100%. Error bars represent the average ± SEM of three independent experiments, significant differences to control treatment (N = 33, t test, ***P < 0.0001). Effect of overexpression of SlNRC4a CRISPR edited fragment on LeEIX2 mediated defence responses in Nicotiana tabacum: (c) Leaf disks of transiently transformed N. tabacum leaves with SlNRC4aCRISPR-GFP or control (free GFP) as indicated (24 hr after transformation) were floated on water for ~24 hr and then replaced with luminescence solution containing 1 μg ml−1 of EIX. Luminescence (RLU) was immediately measured. Average ± SEM values of five independent experiments, n = 12 each. Asterisks indicate significant differences with control treatment (two-way analysis of variance, *P < 0.05). (d) Leaf disks of transiently transformed N. tabacum leaves with SlNRC4aCRISPR-GFP or control (free GFP) as indicated (40 hr after transformation) were floated on a 250 mM of sorbitol solution with 1 μg ml−1 of EIX. Ethylene biosynthesis was measured after 4 hr. Ethylene induction was defined as the ΔEthylene (Ethylene+EIX – Ethylene−EIX). Control induction average value was defined as 100%. Error bars represent the average ± SEM of three independent experiments, with asterisks denoting significant differences with control treatment (N = 18, t test, ***P < 0.0001). Effect of overexpression of SlNRC4a CRISPR-edited fragment on LeEIX2 endosomes in N. benthamiana: Leaves transiently expressing LeEIX2-GFP and free mCherry (Control) or SlNRC4aCRISPR-mCherry as indicated, 40 hr after transformation, leaves were treated with EIX (1 μg g−1 tissue) or water (mock) at the petiole and visualized after 15 min. (e) Representative Z-projections were acquired using Zeiss LSM 780 confocal microscope (40× objective, arrowheads indicate endosomes. Scale bar 30 μm). (f) Number of LeEIX2 endosomes was quantified using 3D object counter (Fiji-ImageJ). Error bars represent the average ± SEM of three independent replicates, four images each. Asterisks indicate significant differences from the control (N = 12, t test, *P < 0.05, **P < 0.001, ***P < 0.0001)
