Fig. 4.
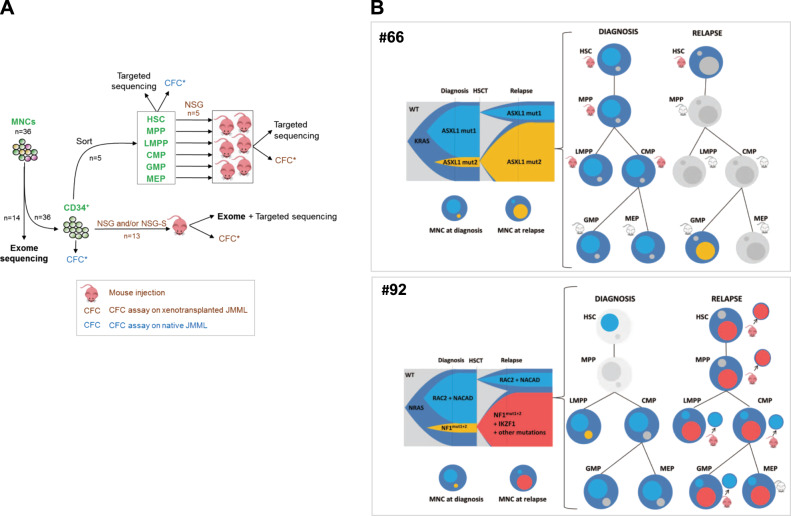
Clonal dynamics with time and across hematopoietic differentiation shows early clonal dominance. a Schematic of experimental procedure followed in order to delineate the origin and clonal architecture of JMML. All CFCs obtained from these experiments on naïve JMML (blue icons) or xenotransplanted JMML (brown icons) were tested for known patient mutations by targeted sequencing. See also Fig. S4. b Clonal architecture of two JMML samples (#66 and #92), as determined by combining whole exome sequencing, deep targeted sequencing, and single-cell derived colony sequencing before and after xenotransplantation. For each patient, a fish plot (left) represents clonal evolution between diagnosis and relapse. In the absence of preleukemia sample allowing to specify the kinetics of clonal emergence, subclones were represented by default as appearing simultaneously. Mutations found in each subclone are indicated (see also Supplementary Table S6). The clonal composition in total JMML mononucleated cells (MNCs) at diagnosis or relapse is also represented in circles. The larger circle represents the founding clone. Smaller circles inside represent subclones of various size and matching colors with the fish plot. Clonal composition and engraftment capacities across hematopoietic differentiation are represented on the right panels. Mutations identified in MNC were screened in sorted fractions before and after xenotransplantation using Sanger sequencing. Mouse icons tag fractions that were injected in NSG and/or NSG-S mice. Red mouse icons indicate successful engraftment whereas gray icons indicate engraftment failure. Patient (#66) KRAS-JMML showing branched evolution with independent acquisition of additional mutations targeting ASXL1. The dominant clone at diagnosis or at relapse was also dominant in corresponding xenografts. Patient (#92) NRAS-JMML showing branched evolution with independent acquisition of additional mutations targeting either NF1 or RAC2. At relapse, exome sequencing performed on the MNC evidenced the gain of an IKZF1 mutation within the clone that became dominant at relapse. Sorted relapse HSC, MPP, and LMPP engrafted in NSG mice and targeted sequencing of individual picked CFC obtained from these mice demonstrated the presence of the dominant relapse clone in HSC and MPP whereas cells retrieved from the LMPP, CMP, and GMP engrafted mice only harbored the mutations of the clone that was dominant at diagnosis but minor at relapse