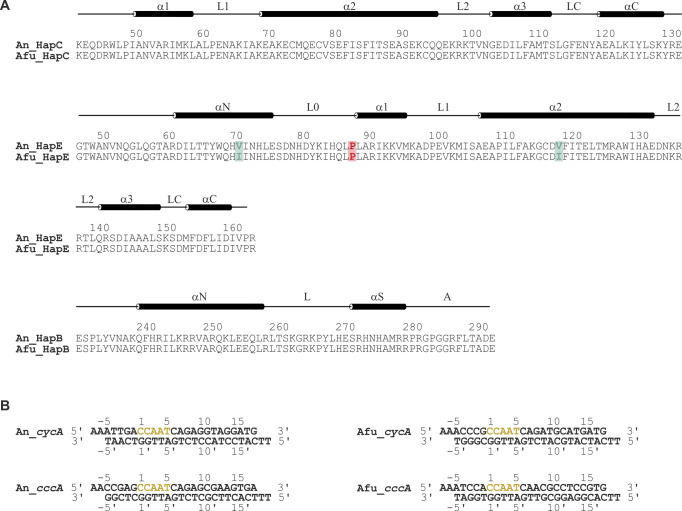
Figure S1. Protein and DNA sequences used for crystallization.
(A) Sequences of the conserved core domains of HapC, HapE, and HapB from Aspergillus nidulans (An) and Aspergillus fumigatus (Afu). Residue numbers are assigned according to the full-length proteins from A. nidulans, derived from the UniProt database. Secondary structures (α: helix) are indicated for the CBC subunits from A. nidulans. Amino acid differences between An_HapE and Afu_HapE are colored green. The site of mutation Pro88 is marked red. (B) DNA duplexes crystallized in complex with CBCs. The CCAAT box is colored yellow. The strand carrying the CCAAT sequence is referred to as Watson strand, and its bases are numbered consecutively, starting with one at the 5′ end of the CCAAT box. Nts upstream of the CCAAT box are assigned negative numbers. Primed bases on the complementary Crick strand are numbered according to their binding partner.