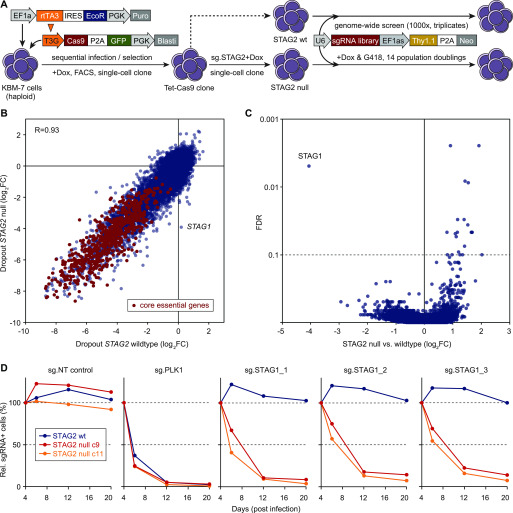
Figure 1. Genome-wide CRISPR screens in isogenic KBM-7 cell lines identify STAG1 as the most selective synthetic-lethal dependency in STAG2-mutant cells.
(A) Schematic of cell line engineering and genome-wide CRISPR screening. Haploid KBM-7 cells were sequentially transduced with indicated lentiviral vectors, Cas9-GFP–expressing cells were single-cell sorted, and derived clones characterized for homogenous and effective CRISPR editing. The selected clone was lentivirally transduced with a validated sgRNA-targeting STAG2, and several subclones were isolated and characterized for STAG2 knockout. One STAG2 knockout clone was selected (“c9”) and screened side-by-side with the parental STAG2–wild-type clone (“B4”) using a second-generation genome-wide sgRNA library. (B) Gene-level dropout effects in wild-type and STAG2-null KBM-7 cells. Shown are log2 fold changes between the end point of triplicate screens and the sgRNA library (analyzed using MAGeCK v0.5.8). A set of previously described generally essential genes (Wang et al, 2019) are highlighted in red. (C) Analysis of differential effects in STAG2-null versus STAG2–wil-type cells (MAGeCK v0.5.8), revealing STAG1 as the most prominent and only significant synthetic lethal interaction. (D) Competitive proliferations assays in STAG2–wild-type (B4, blue lines) and STAG2-mutant (c9, red lines) cell lines used in the screen and in an independent STAG2-mutant clone (c11, orange lines). Cells were transduced with a lentiviral vector co-expressing mCherry and the indicated sgRNAs, and the fraction of mCherry+ cells was monitored over time using flow cytometry.