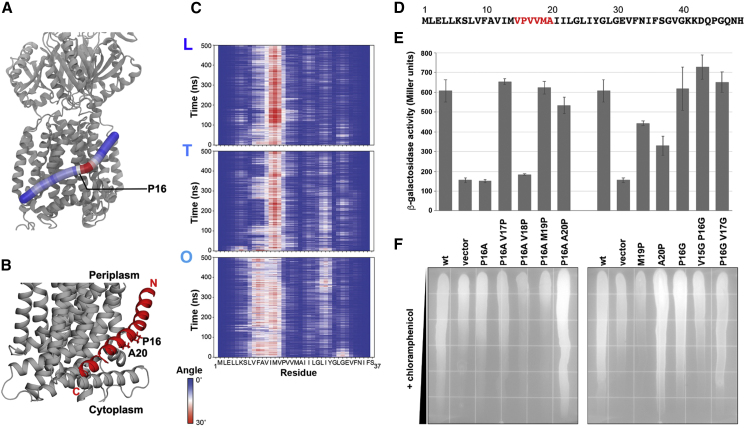
Figure 4.
Impact of AcrZ Curvature on Binding to AcrB
(A) A snapshot of AcrZ bound to the AcrB in the loose conformation. The AcrZ is shown in tube representation and colored based on the bending angle between its residues, from blue (0°) to red (30°), while the AcrB is shown in ribbon representation and colored gray.
(B) A zoom in of AcrZ bound to AcrB. AcrZ is oriented with the N terminus in the periplasm and the C terminus in the cytoplasm. The locations of P16 and A20 are indicated.
(C) The evolution of the bending angle over the course of one of the simulations for all three AcrZ subunits. The conformation of AcrB (L, T, O) to which these AcrZ subunits are bound is shown on top left of each graph.
(D) The sequence of AcrZ with residues mutated indicated in red.
(E) Split adenylate cyclase two-hybrid assays of the interaction between plasmid-encoded T25-AcrB and the empty vector, wild-type AcrZ-T18 or the indicated mutant. T25-AcrB and the AcrZ-T18 indicated were co-expressed in an adenylate cyclase-deficient strain and grown to optical density at 600 nm (OD600) ∼ 1 when cells were harvested for β-galactosidase activity assay. Shown are the average and standard deviation of three experiments. The first and second wild-type (wt) and vector samples are the same.
(F) Exponentially growing cultures of the E. coli ΔacrZ strains carrying the pBAD24 empty vector, wild-type AcrZ, or the indicated AcrZ mutant were applied across chloramphenicol gradient plates to visualize differences in antibiotic sensitivity. The plates were incubated overnight at 37°C and photographed. Shown here is a representative image of an experiment carried out in triplicate.