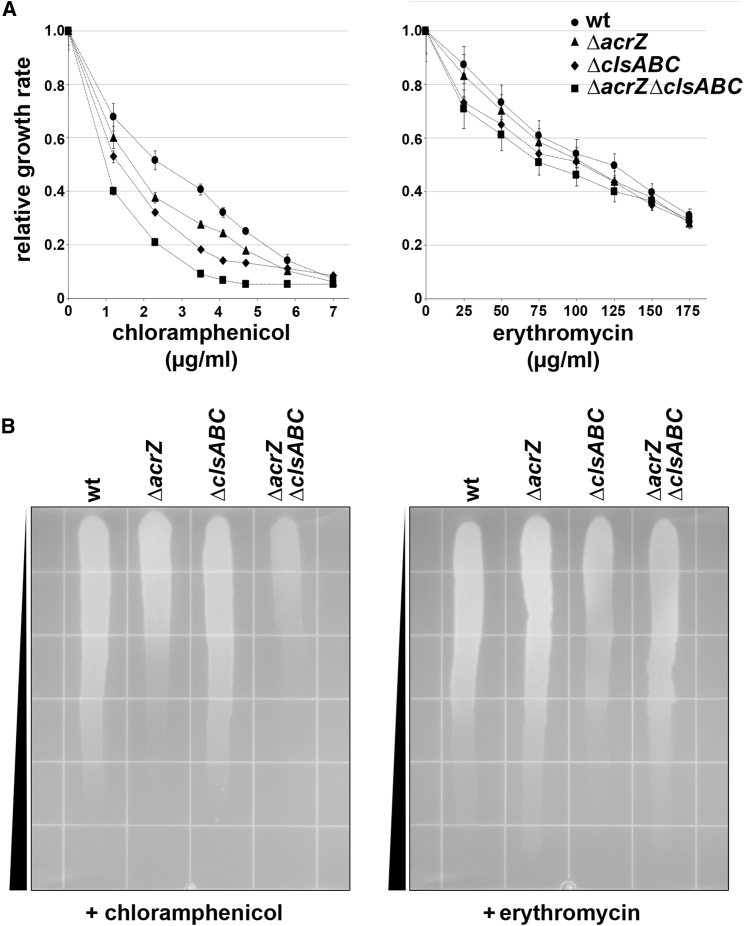
Figure 5.
Cardiolipin Biogenesis and AcrZ Impact on Chloramphenicol Sensitivity
(A) Growth rates of E. coli MG1655 (wild-type parent strain), MG1655 ΔacrZ, MG1655 ΔclsABC::FRT-kan-FRT (cardiolipin-deficient), and MG1655 ΔclsABC::FRT-kan-FRT ΔacrZ (cardiolipin-deficient and ΔacrZ) in the presence of a range of chloramphenicol (0–7 μg mL−1) and erythromycin (0–175 μg mL−1) concentrations were determined relative to the maximum growth rate in the absence of drug. OD660 measurements are presented as mean of triplicate measurements ± standard error of the mean. For the determination of the relative growth rates of the cultures in each of the wells, the exponential phase of the growth curve (as a mean of n = 3 for each culture type) was determined from the linear increase in a log10(OD660) versus time plot. The slope of this section was determined by simple linear regression. Heteroscedasticity-consistent standard errors of the corresponding slope coefficient were calculated. The quality of the fit was significant in all cases (p < 0.05). Next, the relative growth rate was determined as the ratio of the growth rate in the presence of drug over the maximum growth rate in the absence of drug.
(B) Exponentially growing cultures of the above strains were applied across chloramphenicol or erythromycin gradient plates to visualize differences in antibiotic sensitivity. The plates were incubated overnight at 37°C and photographed. Shown is a representative gradient plate image of a biological replicate from experiments carried out for ten different single colonies. The deletion of chromosomal acrZ gave a reproducible but slight increase in resistance to erythromycin in this E. coli strain background, but this mild effect is not observed in a different E. coli strain background (Hobbs et al., 2012).