Figure 6.
Structural Comparison Between Saposin A Disc-Reconstituted AcrB and AcrBZ with Cardiolipin Enrichment
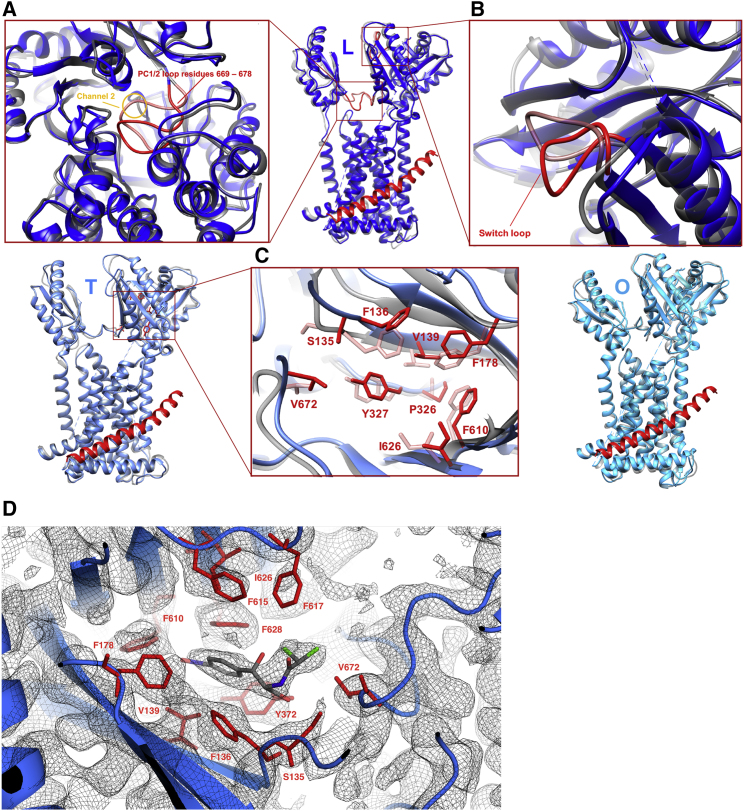
Overlay of protomers in L, T, O of cryo-EM-derived AcrB (in gray; partially transparent) reconstituted in E. coli lipids inside a saposin A disc and AcrBZ (blue variants) reconstituted in E. coli lipids enriched with cardiolipin inside a saposin A disc.
(A) Channel 2 entry is restricted by a loop region of PC1/2 (light red) for substrate entry from the outer leaflet of the inner membrane in case of AcrB in L state but open in the case of the AcrBZ complex in L (dark red).
(B) Once substrate enters the protomer in L, a switch loop (red) allows passage into the deep-binding pocket in AcrB in complex with AcrZ. This loop appears to restrict access in case of the AcrB L protomer in the absence of AcrZ.
(C) Impact on the drug-binding pocket at the site of chloramphenicol binding. Chloramphenicol should be located inside the distal pocket of the AcrB in “tight” conformation. Upon inspection of this area, no discernible chloramphenicol density could be identified, but the antibiotic is predicted to pack against residues P326, Y327, V139, F136, F610, F178, S135, I626, and V672 (red side chains). The residues are mostly on two sets of beta sheets in the porter domain of AcrB, as well as on nearby loops.
(D) Cryo-EM density in the T state pocket for the AcrBZ complex with additional cardiolipin, showing the binding of chloramphenicol (gray) in a discrete conformation, with surrounding residues highlighted. Even though chloramphenicol or minocycline was added to other samples as well, density was not observed in this position for the maps for the AcrBZ and AcrB structures in the natural lipid composition or AcrB with cardiolipin supplement.