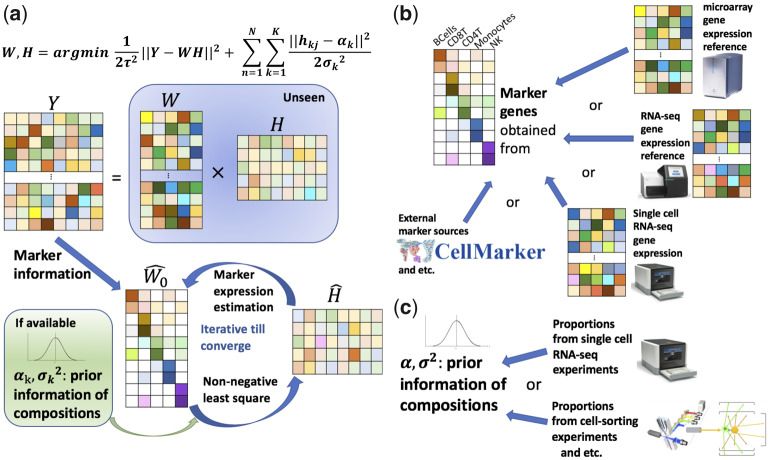
Fig. 1.
A schematic workflow of TOAST. We assume five cell types and eight subjects. (a) TOAST requires a data matrix of tissue expression profiles (Y) and cell-type-specific marker informtion as inputs. Prior information of cell compositions is optional but highly recommended. TOAST adopts an iterative algorithm to solve for pure cell-type profile () and cell compositions (). (b) Cell-type-specific markers can be selected from gene expression reference by microarray or RNA-seq or scRNA-seq, or from existing marker repository, etc. (c) Prior information of cell compositions can be obtained from scRNA-seq or cell-sorting experiments, etc