Figure 2.
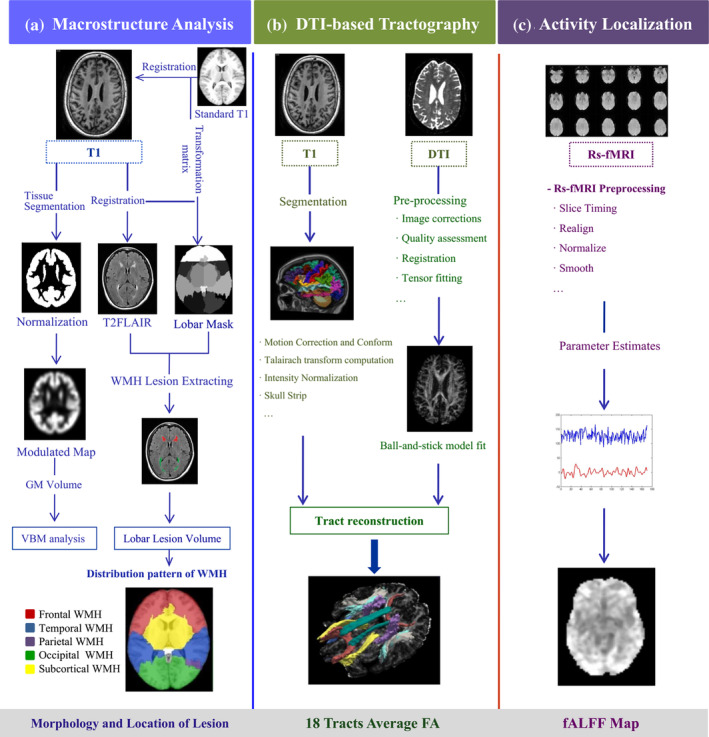
Workflow of multi‐modality imaging pre‐processing. (a) Macrostructure analysis. 3DT1 and T2 FLAIR images were used for the analysis of WMH lesions. 3DT1 images were normalized to the modulated maps following tissue segmentation. We co‐registered the standard atlas to the 3DT1 image of each subject. The 3DT1 image was co‐registered to T2 FLAIR, and the deformation field was obtained for the registration of individual cerebral masks to T2 FLAIR images. The final cerebral masks in the subject's T2 FLAIR space were used to extract the WMH volume in five cerebral ROIs through combing with the WMH lesion map; (b) DTI‐based tractography. The cortical parcellation and subcortical segmentation of raw 3DT1 data and pre‐processing of DTIs were implemented. The probabilistic distribution of the pathways was reconstructed by combining the tracts atlas and labeled structure segmentation data from each subject in the native diffusion space; (c) Activity localization. Rs‐fMRI was used to obtain the fALFF values, and the coefficient map of each subject was normalized to z‐scores across the whole brain after pre‐processing. DTI, diffusion tensor imaging; fALFF, fractional amplitude of low‐frequency fluctuations; GM, gray matter; Rs‐fMRI, resting‐state functional magnetic resonance imaging; WMH, white matter hyperintensity
