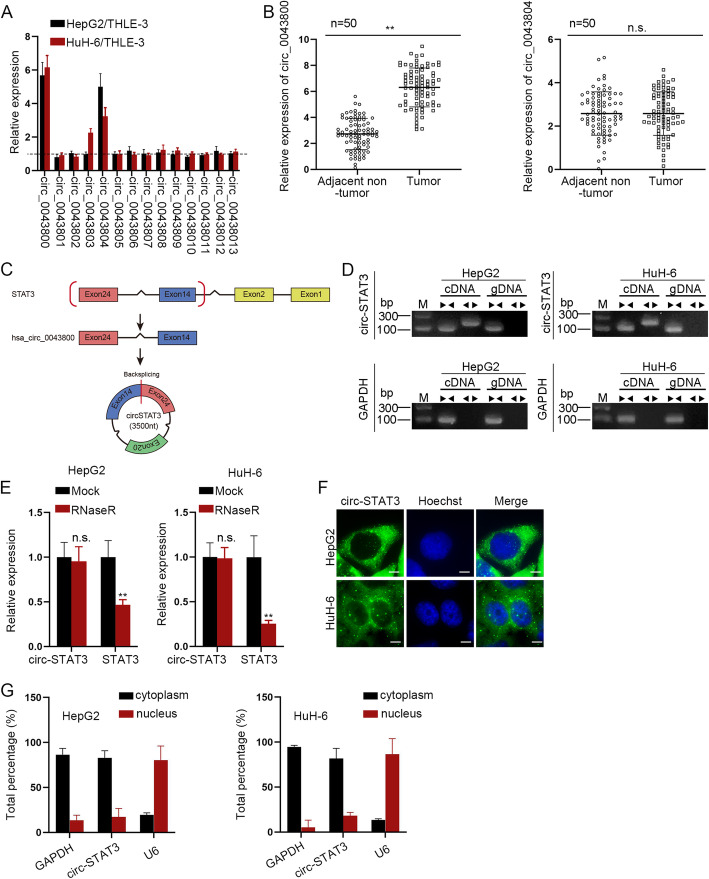
Fig. 1.
Circ_0043800 was up-regulated in HB tissues and cells. a qRT-PCR was applied to measure the expression level of 10 candidate circRNAs in HepG2 and HuH-6 cells (they were referred as HB cells in our study for convenience) compared to control cells of THLE-3. Student’s t-test. b qRT-PCR detected relative expression of circ_0043800 and circ_0043804 in HB tissues and adjacent non-tumor tissues. Student’s t-test. c Schematic diagram of the splicing pattern of circ_0043800 and Sanger sequencing of circ_0043800. d The existence of circ-STAT3 in HB cells was validated by qRT-PCR. e Relative expression of circ-STAT3 and STAT3 in HB cells under the treatment of RNaseR. Student’s t-test. f-g FISH (scale bar: 10 μm) and subcellular fraction assay revealed subcellular location of circ-STAT3. **P < 0.01. The symbol “n.s.” indicates no significance