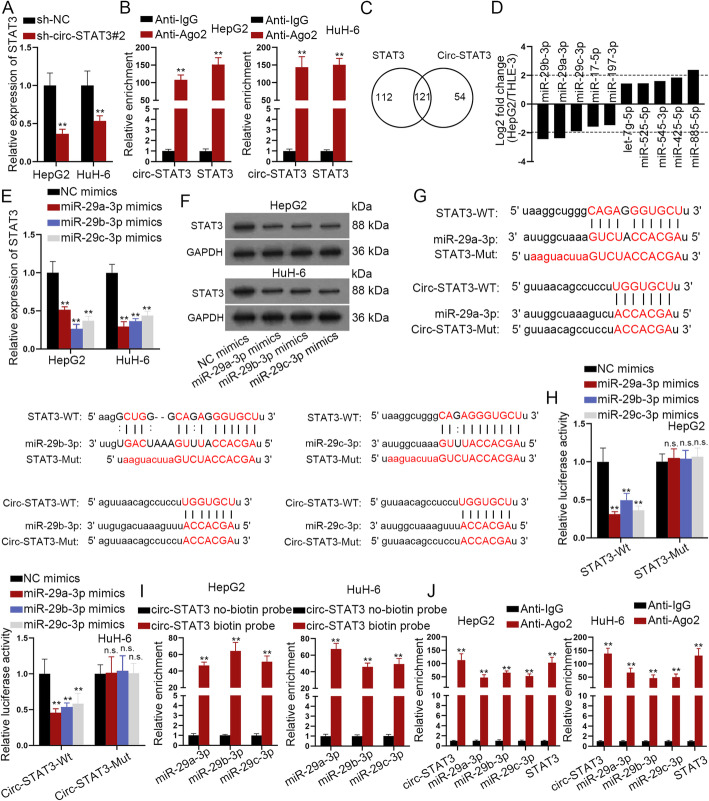
Fig. 3.
Circ-STAT3 sponged miR-29a/b/c-3p to up-regulate STAT3. a Influence of circ-STAT3 depletion on STAT3 expression was evaluated by qRT-PCR analysis. Student’s t-test. b RIP assay revealed enrichment of circ-STAT3 and STAT3 pulled down by anti-IgG and anti-Ago2. Student’s t-test. c Venn diagram revealed the number of miRNAs binding both circ-STAT3 and STAT3 based on starBase database.
d The top 10 significantly abnormally expressed miRNAs in HepG2 cells normalized to control cells. e-f qRT-PCR and western blot examined influence of miR-29a/b/c-3p overexpression on expression of STAT3. One-way ANOVA. g Binding sites of wild type/mutant circ-STAT3/STAT3 and miR-29a/b/c-3p. h Luciferase reporter assay revealed luciferase activity of wild and mutant circ-STAT3/STAT3 (3 different mutated circ-STAT3/STAT3 with different mutations for miR-29a/b/c-3p) by miR-29a/b/c-3p mimics. One-way ANOVA. i RNA pull down assay revealed enrichment of miR-29a/b/c-3p pulled down by biotinylated circ-STAT3 and non-biotinylated circ-STAT3. Student’s t-test. j RIP assay revealed enrichment of circ-STAT3, miR-29a/b/c-3p and STAT3 in anti-IgG and anti-Ago2. Student’s t-test. **P < 0.01. The symbol “n.s.” indicates no significance.