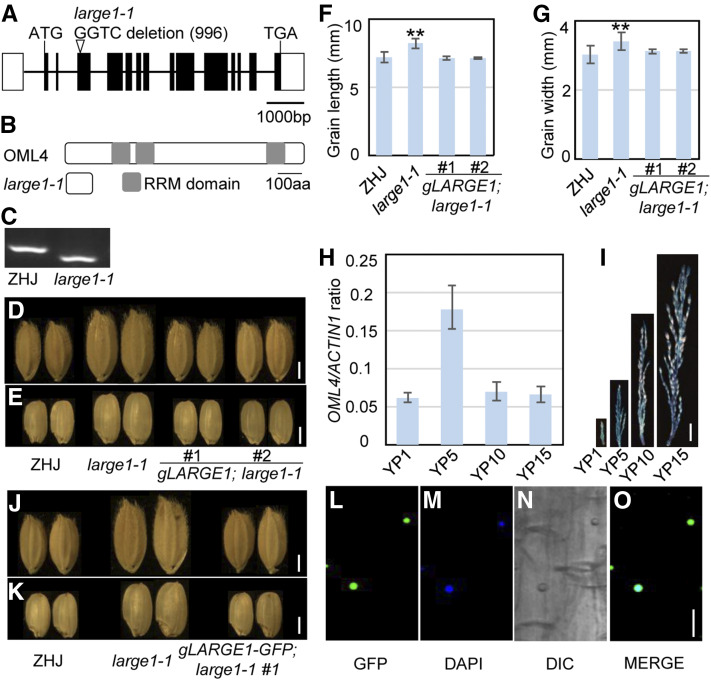
Figure 3.
LARGE1 Encodes the Mei2-Like Protein OML4.
(A) LARGE1/OML4 gene structure. The coding sequence is shown as the black box, and introns are indicated using black lines. ATG and TGA represent the start codon and the stop codon, respectively.
(B) OML4 and mutated protein encoded by large1. The OML4 protein contains three RRM domains. The mutation results in a premature termination codon in OML4, causing a truncated protein.
(C) Derived cleaved-amplified polymorphic sequence marker was developed according to the large1-1 mutation. The PCR products were digested by the restriction enzyme HphI.
(D) and (E) Mature paddy (D) and brown (E) rice grains of ZHJ, large1-1, gLARGE1;large1-1 #1, and gLARGE1;large1-1 #2.
(F) and (G) Grain length (F) and width (G) of ZHJ, large1-1, gLARGE1;large1-1 #1, and gLARGE1;large1-1 #2. Asterisks indicate significant differences between ZHJ and large1-1. **, P < 0.01 compared with the wild type by Student’s t test.
(H) Relative OML4 gene expression in young panicles of 1 cm (YP1) to 15 cm (YP15) in ZHJ. RT-qPCR was used to measure expression levels of OML4 in panicles. Values are given as mean ± sd. Three biological replicates were used (n = 3).
(I) OML4 expression activity was monitored by proOML4:GUS transgene expression. Histochemical analysis of GUS activity in panicles at different developmental stages.
(J) and (K) Mature paddy (J) and brown (K) rice grains of ZHJ, large1-1, and gLARGE1-GFP;large1-1 #1.
(L) to (O) Subcellular location of OML4-GFP in gLARGE1-GFP;large1-1 #1 root cells. GFP fluorescence of GFP-OML4 (L), 4′,6-diamidino-2-phenylindole staining (DAPI; see [M]), differential interference contrast (DIC; see [N]), and merged (O) images are shown.
Bar in (D), (E), (J), and (K) = 2 mm; bar in (I) = 1 cm; bar in (L) to (O) = 10 µm.