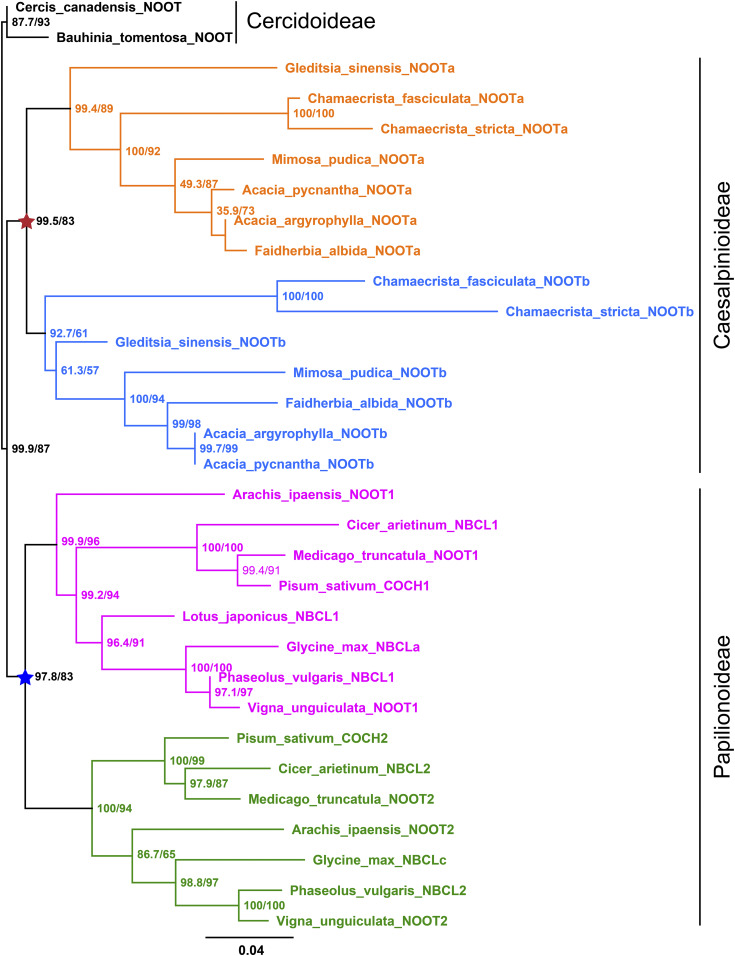
Figure 6.
Maximum Likelihood Tree of Legume NOOT/NBCL Proteins.
The NOOT/NBCL proteins are present in three legume subfamilies, with Cercidoideae representing the most basal one. The Cercidoideae family members for which sequence data are available, Cercis canadensis and Bauhinia tomentosa, do not form nodules and contain a single NOOT/NBCL gene. Except for Lotus japonicus and Glycine max, all other legumes (subfamilies Papilionoideae and Caesalpinioideae) for which sequence data are available have two NOOT/NBCL genes that are grouped into four major clades, with two clustered clades containing Papilionoideae NOOT/NBCL genes (indicated in pink and green) and the other clades containing Caesalpinioideae NOOT/NBCL genes (indicated in orange and blue). This suggests that the NOOT/NBCL gene was independently duplicated in the common ancestor of Papilionoideae (blue star) and in that of Caesalpinioideae (red star). Numbers at the nodes indicate approximate Bayes support (%)/ultrafast bootstrap support (%). Ultrafast bootstrap approximation values are based on 5000 replicates. The protein sequences used in this analysis are summarized in Supplemental Table 1. For convenience, previously unnamed NOOT/NBCL proteins were all named NOOT in this analysis. Glycine max NBCLb (Glyma19g131000.1) is a truncated protein and was therefore not included in this analysis. C. canadensis and B. tomentosa from the Cercidoideae subfamily are the two most basal legumes in the analysis; therefore, C. canadensis NOOT and B. tomentosa NOOT were used as the outgroup. Branch lengths were scaled (see the bottom) and represent the number of substitutions per site. Note that the asterisks are shown in the same color and position as in Supplemental Figure 1.