Tre6P synthesis by TPS1 is essential for embryogenesis and postembryonic growth in Arabidopsis, and appropriate Suc signaling by Tre6P is dependent on the noncatalytic domains of TPS1.
Abstract
In Arabidopsis (Arabidopsis thaliana), TREHALOSE-6-PHOSPHATE SYNTHASE1 (TPS1) catalyzes the synthesis of the sucrose-signaling metabolite trehalose 6-phosphate (Tre6P) and is essential for embryogenesis and normal postembryonic growth and development. To understand its molecular functions, we transformed the embryo-lethal tps1-1 null mutant with various forms of TPS1 and with a heterologous TPS (OtsA) from Escherichia coli, under the control of the TPS1 promoter, and tested for complementation. TPS1 protein localized predominantly in the phloem-loading zone and guard cells in leaves, root vasculature, and shoot apical meristem, implicating it in both local and systemic signaling of Suc status. The protein is targeted mainly to the nucleus. Restoring Tre6P synthesis was both necessary and sufficient to rescue the tps1-1 mutant through embryogenesis. However, postembryonic growth and the sucrose-Tre6P relationship were disrupted in some complementation lines. A point mutation (A119W) in the catalytic domain or truncating the C-terminal domain of TPS1 severely compromised growth. Despite having high Tre6P levels, these plants never flowered, possibly because Tre6P signaling was disrupted by two unidentified disaccharide-monophosphates that appeared in these plants. The noncatalytic domains of TPS1 ensure its targeting to the correct subcellular compartment and its catalytic fidelity and are required for appropriate signaling of Suc status by Tre6P.
INTRODUCTION
Trehalose 6-phosphate (Tre6P) is a potent signaling molecule in plants. The enzyme responsible for its biosynthesis, trehalose-6-phosphate synthase (TPS; EC 2.4.1.15), is essential for plant viability (Eastmond et al., 2002; van Dijken et al., 2004; Gómez et al., 2010). Tre6P is the phosphorylated intermediate in the two-step pathway of trehalose biosynthesis (Cabib and Leloir, 1958); it is synthesized from UDP-glucose and glucose 6-phosphate (Glc6P) by TPS, then dephosphorylated to trehalose by trehalose-6-phosphate phosphatase (TPP; EC 3.1.3.12). The importance, indeed the very existence, of this pathway in flowering plants has only emerged over the last 20 years. The unexpected finding of genes encoding functional TPS (TPS1; Blázquez et al., 1998) and TPP (TPPA and TPPB; Vogel et al., 1998) enzymes in Arabidopsis (Arabidopsis thaliana) was followed by the discovery that loss-of-function mutants of TPS1 are nonviable, with homozygous tps1 embryos failing to complete embryogenesis, becoming arrested at the torpedo stage (Eastmond et al., 2002). Arrested embryos have fewer cells, indicating defective cell division, as well as abnormalities in their cell wall structure and starch content (Eastmond et al., 2002; Gómez et al., 2006). However, the underlying cause of the embryo-arrest phenotype has not been established. Viable tps1 seeds can be obtained by dexamethasone-inducible expression of TPS1 during seed development (van Dijken et al., 2004) or by embryo-specific expression of TPS1 under the control of the ABSCISIC ACID INSENSITIVE3 promoter (Gómez et al., 2010). Nonetheless, the resulting tps1 plants are severely dwarfed and either do not flower or flower very late (van Dijken et al., 2004; Gómez et al., 2010; Wahl et al., 2013), showing that a functional TPS1 is needed for normal growth and development throughout the plant life cycle.
A key breakthrough came with the discovery that increasing or decreasing the levels of Tre6P in Arabidopsis, by constitutive expression of Escherichia coli TPS (35Spro:otsA) or TPP (35Spro:otsB), led to profound but opposite changes in plant morphology (Schluepmann et al., 2003). 35Spro:otsA plants had small leaves, precocious flowering, and a bushy phenotype, whereas 35Spro:otsB plants had large leaves, delayed flowering, and only one or a few shoot branches. These experiments revealed the potent influence of Tre6P on plant growth and development. The levels of Tre6P in plants were found to be highly correlated with Suc (Lunn et al., 2006), which led to the proposal that Tre6P functions as a signal of Suc status. Elaborating on this idea, our current sucrose-Tre6P nexus model postulates that Tre6P is not only a signal of Suc status but also a negative feedback regulator of Suc levels, acting in a way that is reminiscent of the homeostatic control of blood glucose levels in animals by insulin (Yadav et al., 2014). Suc dominates the metabolism of flowering plants; it is the major product of photosynthesis, the most common transport sugar, and the main source of carbon and energy in growing sink organs (Lunn, 2016). This dominance may explain why Tre6P, acting as a signal and regulator of Suc levels, can exert such a far-reaching influence on plant growth and development (Figueroa and Lunn, 2016). In Suc-producing source leaves, Tre6P regulates Suc levels by modulating photoassimilate partitioning during the day (Figueroa et al., 2016) and the mobilization of transitory starch reserves at night (Martins et al., 2013; Dos Anjos et al., 2018). In Suc-consuming sink organs, Tre6P regulates the utilization of Suc for growth and accumulation of storage products, acting, at least in part, via the inhibition of SUCROSE-NON-FERMENTING1-RELATED KINASE1 (Zhang et al., 2009; Nunes et al., 2013; Zhai et al., 2018). Although the reciprocal regulation of Suc and Tre6P appears to operate in somewhat different ways in source and sink tissues, the sucrose-Tre6P nexus model offers a unifying concept for the fundamental role of Tre6P in plants (Figueroa and Lunn, 2016).
The recognition of Tre6P as a potent regulator of plant growth and development suggested that the arrest of tps1 embryos at the torpedo stage might be due to impaired synthesis of Tre6P. However, in common with other flowering plants, Arabidopsis has a large family of TPS genes encoding TPS or TPS-like proteins (Leyman et al., 2001; Avonce et al., 2006; Lunn, 2007). The 11 TPS genes in Arabidopsis form two distinct clades: class I (TPS1 to TPS4) and class II (TPS5 to TPS11). With the exception of TPS3, which is most likely a pseudogene, only class I TPS genes have been reproducibly shown to encode catalytically active TPS enzymes (Blázquez et al., 1998; Van Dijck et al., 2002; Vandesteene et al., 2010; Delorge et al., 2015). At the moment, the functions of the noncatalytic class II TPS-like proteins are poorly understood (Harthill et al., 2006; Ramon et al., 2009; Lunn et al., 2014). TPS2 and TPS4 are almost exclusively expressed in developing seeds (Vandesteene et al., 2010). However, despite being catalytically active (Delorge et al., 2015), the encoded enzymes do not compensate for the loss of TPS1 in developing tps1 mutant seeds. It is unclear whether this is due to differences in their spatiotemporal expression patterns (i.e., TPS2 and TPS4 are not expressed in the right place at the right time in developing seeds to substitute for TPS1) or if the TPS1 protein has unique properties that the other two class I TPS isoforms lack (reviewed in Figueroa and Lunn, 2016). In this context, it is worth noting that two TPP genes in maize (Zea mays), RAMOSA3 and ZmTPP4, were recently found to encode catalytically active TPP enzymes that also have noncatalytic functions in regulating inflorescence branching (Satoh-Nagasawa et al., 2006; Claeys et al., 2019), setting a precedent for enzymes in this pathway to have “moonlighting” functions.
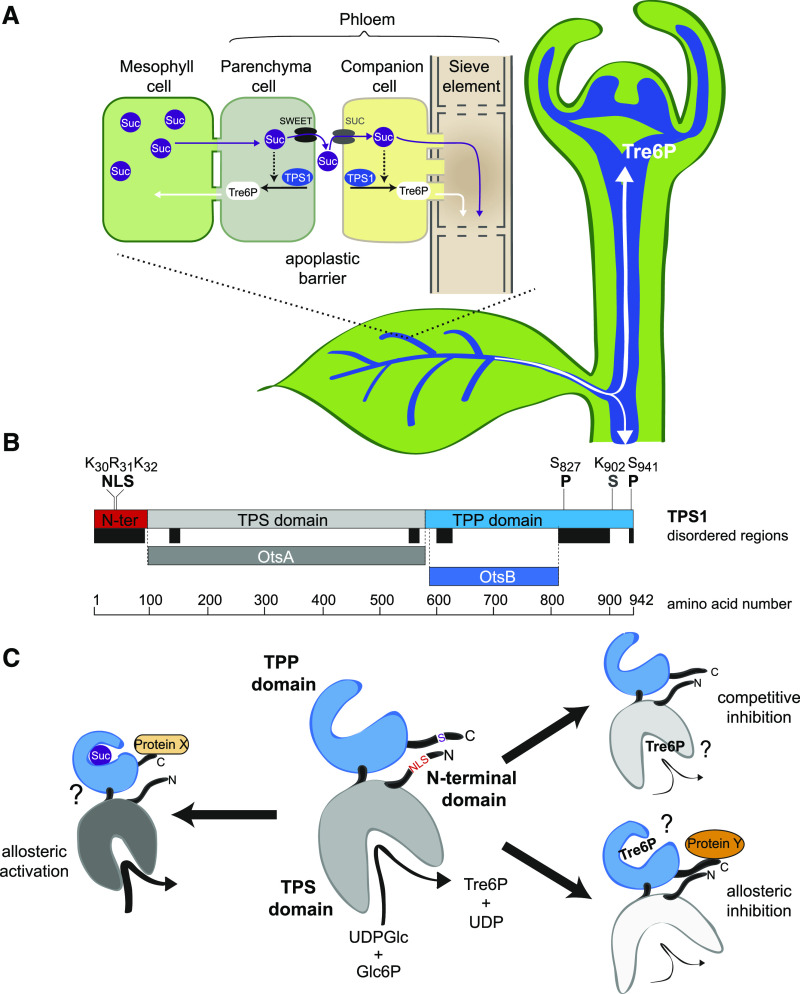
The Arabidopsis TPS1 protein has three distinct domains: (1) an N-terminal domain that includes a putative autoinhibitory Leu/Arg-rich motif; (2) a glucosyltransferase domain containing the catalytic site; and (3) a TPP-like C-terminal domain of unknown function (Figure 1B; Van Dijck et al., 2002; Lunn, 2007). The other two catalytically active isoforms, TPS2 and TPS4, also have glucosyltransferase and C-terminal TPP-like domains, but they lack an equivalent of the TPS1 N-terminal domain. Curiously, such truncated class I TPS proteins are found only in the Brassicaceae, the significance of which is unknown (Lunn, 2007). As the predominant Tre6P-synthesizing enzyme in Arabidopsis, TPS1 might be expected to play a major role in determining the levels of Tre6P, but how the enzyme is regulated and contributes to the sucrose-Tre6P nexus is poorly understood (Yadav et al., 2014).
Figure 1.
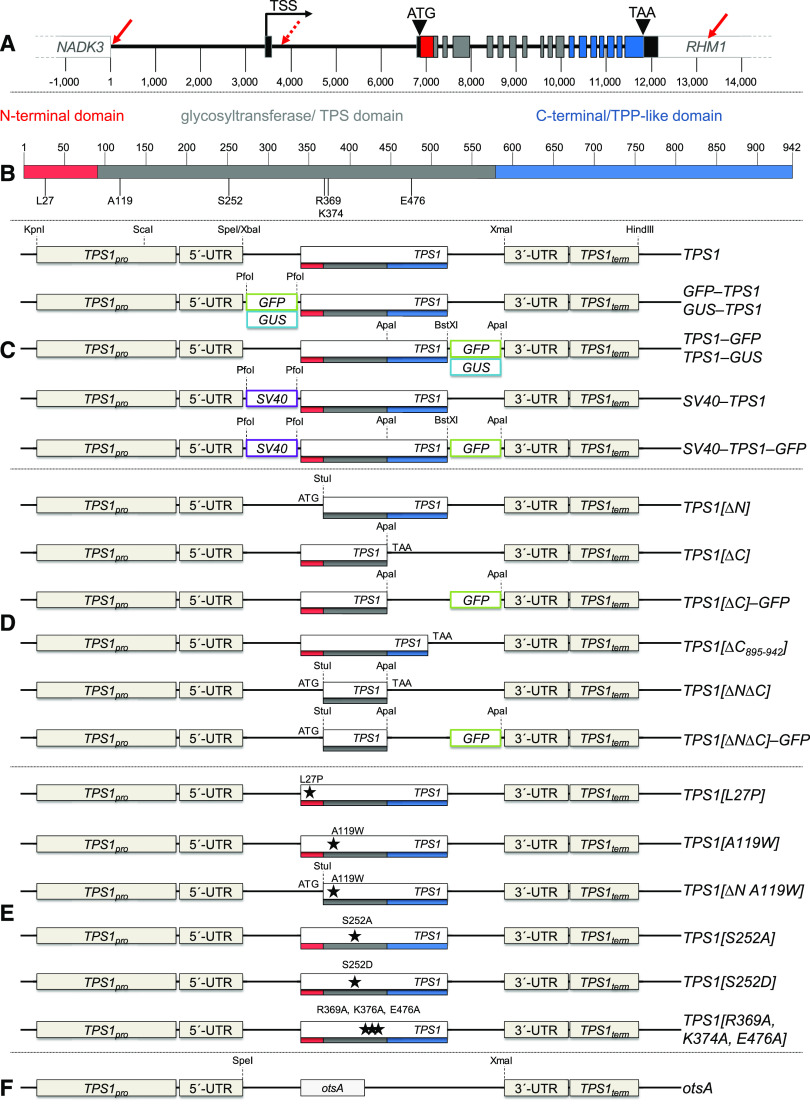
Constructs for Complementation of the Arabidopsis tps1-1 Knockout Mutant by Tagged, Truncated, or Mutated Versions of TPS1 or a Heterologous TPS.
(A) TPS1 constructs are derived from the TPS1 (At1g78580) genomic locus, including the native promoter (TPS1pro) and terminator (TPS1term) regions. The TPS1 gene contains 18 exons, represented by bars that are color-coded: 5′- and 3′-UTRs (black); exons encoding the N-terminal (red), catalytic (gray), and C-terminal (blue) domains. Solid red arrows, 5′- and 3′-ends of the TPS1 genomic region used in this study; dashed red arrow, 5′-end of the presumed promoter region used in previous studies; TSS, transcription start site.
(B) TPS1 functional domains: N-terminal domain (red), catalytic (TPS) glucosyltransferase domain (gray), and C-terminal domain with similarity to TPP (blue).
(C) Constructs encoding full-length versions of TPS1: wild-type (TPS1) and fusion proteins tagged with GFP (green box), GUS (blue box), or the SV40 NLS (purple box).
(D) Constructs encoding truncated forms of TPS1 with deletions of the N-terminal domain (TPS1[ΔN]), all or part of the C-terminal domain (TPS1[ΔC] and TPS1[ΔC895-942]), or both (TPS1[ΔNΔC]).
(E) Constructs encoding mutated forms of TPS1 with single or triple point mutations in the N-terminal or catalytic domain.
(F) Construct encoding the heterologous TPS from E. coli (otsA) under the control of the TPS1 and other potential gene regulatory elements. Dashed lines indicate restriction sites used for cloning.
Note that the various elements of the gene constructs represented in (C) to (F) are not drawn to scale.
Surprisingly little is known about the cellular and subcellular localizations of TPS1, given that it is essential for plant viability. Microarray analysis has shown that TPS1 is expressed in all major tissues—leaves, roots, and reproductive organs (Supplemental Figure 1; Schmid et al., 2005)—but less is known about its expression in specific cell types. TPS1 transcripts are relatively abundant in bundle sheath cells in Arabidopsis, based on ribosome pull-down experiments (Aubry et al., 2014). RNA in situ hybridization also demonstrated transcript accumulation in the protovasculature of developing leaves and in the shoot apical meristem (SAM; Wahl et al., 2013). TPS1 protein was detected in a proteomic analysis of Arabidopsis guard cells (Zhao et al., 2008). In cucumber (Cucumis sativus), the orthologous CsTPS1 protein was found in phloem sap exudates (Hu et al., 2016). In several previous studies in Arabidopsis, genomic regions of 2 to 3 kb upstream of the TPS1 protein-coding region were used to drive the expression of a TPS1 cDNA or bacterial otsA in tps1 mutant complementation experiments or the expression of a GUS reporter gene for promoter analysis (Schluepmann et al., 2003; van Dijken et al., 2004; Gómez et al., 2010; Vandesteene et al., 2010). However, the presumed promoter region used in those studies was chosen on the basis of an incompletely annotated version of the TPS1 locus (At1g78580) that missed the first exon, which only encodes part of the 5′-untranslated region (UTR) and lies 3.2 kb upstream of the first protein-coding exon (Figure 1A). Thus, the above-mentioned studies used a section of intron 1 and part of the 5′-UTR (exon 2) to drive TPS or GUS gene expression in their complementation and promoter analysis constructs, so the introduced genes almost certainly had spatiotemporal expression patterns that were different from the endogenous TPS1 gene. There are also conflicting reports on the subcellular compartmentation of TPS1. The SUBA4 database (Hooper et al., 2017; suba.live) shows no consensus from sequence-based prediction tools for the intracellular location of TPS1 but notes that TPS1 has been experimentally detected in plasma membrane-enriched fractions (Mitra et al., 2009), although the protein has no obvious transmembrane domains. By contrast, transient expression of a construct encoding GFP-tagged TPS1 in Arabidopsis protoplasts indicated that the protein is predominantly cytosolic, although some is located in the nucleus (Vandesteene et al., 2010).
In this study, we explored the elements and functional features of Arabidopsis TPS1 to understand why this enzyme, from a rather obscure branch of sugar metabolism, has the power to determine plant fate. We generated complementation lines of the Arabidopsis tps1-1 null mutant with GUS- or GFP-tagged versions of the TPS1 protein, expressed under the control of the endogenous TPS1 promoter and other potential gene regulatory elements, to elucidate the tissue- and cell-specific accumulation pattern of the TPS1 protein and its subcellular compartmentation. We used a similar strategy to investigate the functions of individual domains and residues within TPS1 and to identify potential moonlighting functions by attempting to complement the tps1-1 mutant with various truncated or mutated versions of TPS1 or with a heterologous TPS (OtsA) enzyme from E. coli. Our results reveal that TPS1, and by inference Tre6P synthesis, predominantly localizes to guard cells and around the phloem-loading zone in source leaves, a strategically important site in source-sink relations and for systemic signaling. We show that loss of the Tre6P-synthesizing capacity of TPS1 is the fundamental reason why tps1-1 mutant embryos arrest at the torpedo stage and is a major, but not necessarily the only, factor underlying their postembryonic growth defects. We observed that the N-terminal domain of TPS1 regulates its distribution between nucleus and cytosol and that TPS1 misbehaves if the C-terminal TPP-like domain is missing, with catastrophic consequences for the plant.
RESULTS
Constructs for the Expression of Wild-Type and Mutated Forms of TPS Proteins
The gene constructs used to test for complementation of the tps1-1 mutant were all derived from the Arabidopsis Columbia-0 (Col-0) TPS1 genomic sequence, based on the current annotation of the TPS1 locus (At1g78580) in the Araport11 database (https://www.araport.org/data/araport11; Cheng et al., 2017). All constructs (Figures 1C to 1F) included the entire intragenic region (∼3.5 kb) between the transcription start site of TPS1 and the end of the coding region of the neighboring NADH KINASE3 (NADK3; At1g78590) gene. This upstream region is presumed to contain the true TPS1 promoter region (TPS1pro). In addition, the constructs contained the 5′-UTR (exon 1 + intron 1 + part of exon 2) and a large region (∼1000 bp) downstream of the translation stop codon containing the 3′-UTR (part of exon 18) and terminator region (TPS1term), including part of the neighboring RHAMNOSE BIOSYNTHESIS1 (RHM1; At1g78570) gene where the polyadenylation site of TPS1 is located. Constructs for the expression of full-length versions of the TPS1 gene (native or mutated) also contained all of the endogenous TPS1 intronic sequences, and introns were retained in the protein-coding regions that were included for the expression of truncated forms of TPS1 (Figure 1A). By using the endogenous TPS1 promoter and other potential gene regulatory elements, we expected the introduced transgenes to have the same spatiotemporal expression pattern and expression level as the wild-type TPS1 gene. As a positive control for complementation, we used the full-length TPS1 gene with no modifications (TPS1; Figure 1C).
The first group of constructs encoded full-length TPS1 proteins tagged at the N or C terminus with either GFP (GFP-TPS1 and TPS1-GFP) or GUS (GUS-TPS1 and TPS1-GUS) to determine the cellular and subcellular localizations of TPS1 (Figure 1C). To investigate the potential role of TPS1 in the nucleus (Vandesteene et al., 2010), we also generated constructs encoding TPS1 (with or without the C-terminal GFP) with a strong nuclear localization signal (NLS) from the Simian Vacuolating Virus 40 (SV40) added at the N terminus (SV40-TPS1 and SV40-TPS1-GFP; Figure 1C). The second group of constructs (Figure 1D) encoded truncated TPS1 proteins lacking either the N-terminal (TPS1[∆N]) or C-terminal (TPS1[∆C]) domain or both (TPS1[∆N∆C]). We also made GFP-tagged variants of the latter two (TPS1[∆C]-GFP and TPS1[∆N∆C]-GFP). The third group of constructs (Figure 1E) encoded full-length TPS1 proteins with substitutions of one or more amino acid residues: (1) L27P, expected to abolish the autoinhibitory function (Van Dijck et al., 2002); (2) A119W, expected to compromise catalytic activity (Vandesteene et al., 2010); (3) S252A, removing a putative phosphorylation site (Glinski and Weckwerth, 2005); (4) S252D, to mimic a constitutively phosphorylated Ser-252; and (5) R369A/K374A/E476A, targeting a triad of active site residues that are highly conserved in TPS and related glucosyltransferases and expected to abolish catalytic activity (Wu et al., 2015). The final construct contained the E. coli otsA gene (Figure 1F), encoding a simple form of TPS (OtsA) with only a single catalytic glucosyltransferase domain and no obvious regulatory potential if expressed in plants. This construct was designed to test whether the tps1-1 mutant could be complemented by replacing only the Tre6P-synthesizing activity of TPS1.
Complementation of the Arabidopsis tps1-1 Null Mutant
All constructs carried the neomycin phosphotransferase II (nptII) gene in the T-DNA and conferred kanamycin resistance. They were introduced into glufosinate-resistant heterozygous TPS1/tps1-1 plants (Eastmond et al., 2002) by floral dip transformation (Clough and Bent, 1998). We selected primary transformants for dual resistance to kanamycin and glufosinate, allowed them to self, and screened the T2 progeny for homozygosity of the tps1-1 locus by genomic PCR (Supplemental Figures 2A and 3A). Immunoblotting with antibodies raised against TPS1 (α-TPS1; Yadav et al., 2014) confirmed the absence of the endogenous full-length (106-kD) TPS1 protein and, where possible, the expression of introduced TPS1 proteins.
With a single exception, we obtained at least one viable line that was homozygous for the tps1-1 mutant allele (tps1-1/tps1-1) for each construct, demonstrating complementation of the tps1-1 null mutation during embryogenesis. The only exception was the mutated catalytic triad construct: TPS1[R369A,K374A,E476A]. We screened the T2 progeny from two independent lines for this construct by genomic PCR, after selection for plants resistant to both kanamycin and glufosinate; all 36 out of 36 plants from one line and all 60 out of 60 plants from the other line were heterozygous (TPS1/tps1-1) for the tps1-1 allele. The failure to recover any TPS1[R369A,K374A,E476A] plants with a homozygous tps1-1/tps1-1 background indicated that the catalytically inactive form of TPS1 encoded by this construct was unable to rescue the tps1-1 mutant through embryogenesis. For constructs that gave rise to an aberrant phenotype, we confirmed the phenotype in at least two independent lines or in independent lines expressing the same modified TPS1 protein with or without a GFP tag (e.g., TPS1[ΔNΔC]-GFP and TPS1[ΔNΔC]). The only exception was TPS1[ΔN], for which we obtained a single line that showed only a very mild phenotype. Supplemental Figure 4 summarizes all analyzed tps1-1 complementation lines.
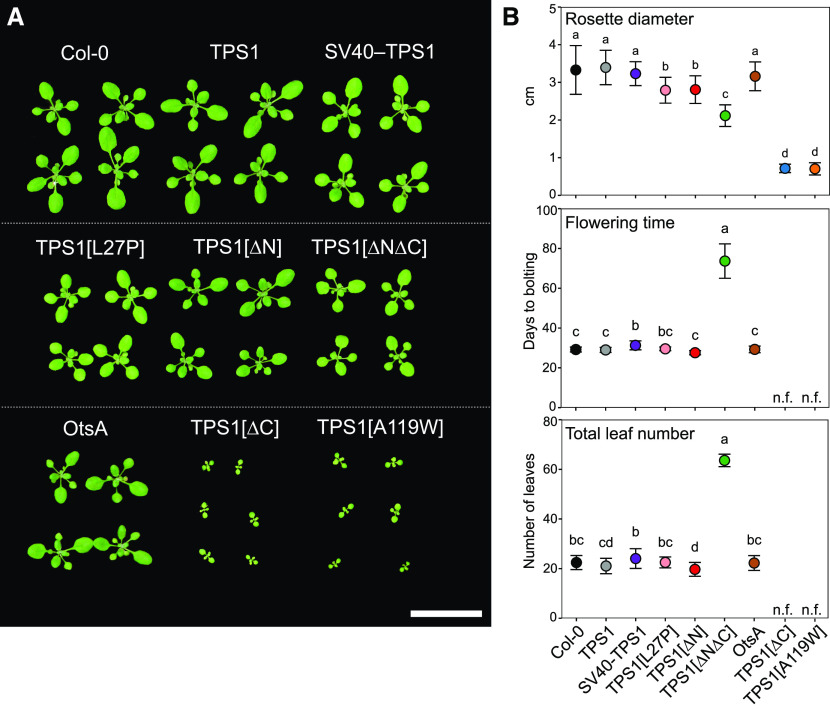
Two independent TPS1 lines, complemented with the wild-type TPS1 gene, were indistinguishable from wild-type plants, with no significant differences in rosette diameter (Figure 2; Supplemental Figure 4), flowering time (Figures 2B and 3), primary root growth (Supplemental Figure 5), or shoot branching (Supplemental Figure 6). This indicated that the TPS1 genomic construct, from which all other constructs were derived, fully complemented the tps1-1 mutant. Complementation lines with constructs expressing GUS- or GFP-tagged versions of the full-length TPS1 gene also showed no obvious differences from wild-type Col-0 or TPS1-complemented plants (Supplemental Figures 2C and 4). The SV40-TPS1 (±GFP) and OtsA lines had wild type-like rosettes (Supplemental Figure 4) and flowered at the same time as Col-0 and TPS1 plants (Figure 2B), but they had much shorter primary roots (Supplemental Figure 5). The TPS1[L27P] lines had slightly smaller rosettes, but flowering time and root growth were unaffected (Figure 2; Supplemental Figure 5). The TPS[S252A] and TPS1[S252D] lines had no obvious phenotypes (Supplemental Figure 4), indicating that changes in the phosphorylation status of the putative Ser-252 phosphorylation site (Glinski and Weckwerth, 2005) have little or no impact on TPS1 protein function in vivo. In contrast, the TPS1[A119W] lines were severely dwarfed and never flowered, even when grown for 5 months under inductive long-day conditions (Figure 2; Supplemental Figure 6).
Figure 2.
Morphological Phenotypes of tps1-1 Complementation Lines under Long-Day Conditions.
Wild-type Arabidopsis Col-0 plants and tps1-1 complementation lines were grown in a 16-h photoperiod.
(A) Rosette morphology of plants 17 d after sowing. Bar = 3 cm.
(B) Rosette diameter (top; n = 17 to 30) and flowering time based on days to bolting (middle; n = 15 to 20) or total leaf number (bottom; n = 10 to 20). Data are presented as means ± sd, and n indicates the number of biological replicates. Letters represent significant differences (P ≤ 0.05) based on ANOVA with posthoc LSD testing. n.f., nonflowering 150 d after sowing. Black symbols represent wild-type Col-0. Other symbol colors represent the tps1-1 mutant transformed with constructs shown in Figure 1: TPS1 (line no. 2; gray), SV40-TPS1 (line no. 1; purple); TPS1[L27P] (line no. 5; pink), TPS1[∆N] (red), TPS1[∆N∆C] (green), otsA (line no. 2; brown), TPS1[∆C] (line no. 7; blue), and TPS1[A119W] (line no. 6; orange).
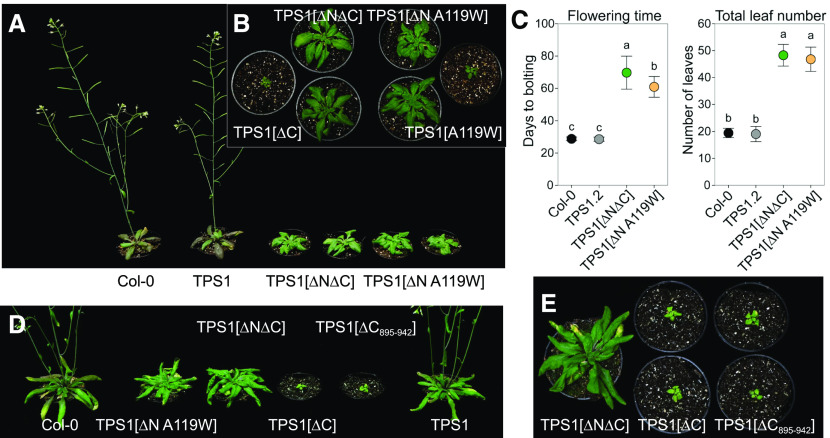
Figure 3.
Morphological Phenotypes of tps1-1 Complementation Lines under Long-Day Conditions.
Wild-type Arabidopsis Col-0 plants and tps1-1 complementation lines were grown in a 16-h photoperiod.
(A) and (B) Shoot morphology of plants 42 d after sowing.
(C) Flowering time based on days to bolting and total leaf number. Data are presented as means ± sd (n = 10 to 14). Letters represent significant differences (P ≤ 0.05) based on ANOVA with posthoc LSD testing. Black symbols represent wild-type Col-0. Other symbol colors represent the tps1-1 mutant transformed with constructs shown in Figure 1: TPS1 (gray), TPS1[ΔN∆C] (green), and TPS1[ΔN A119W] (orange). The TPS1[ΔC], TPS1[ΔC895-942], and TPS1[A119W] lines did not flower.
(D) and (E) Shoot morphology of plants at 45 d after sowing.
Among the lines transformed with constructs expressing truncated forms of TPS1, the TPS1[∆N] line was the most similar to wild-type plants, showing only a small decrease in rosette diameter (Figure 2) and primary root growth (Supplemental Figure 5) but no significant difference in flowering time from wild-type Col-0 (Figure 2B). By contrast, the TPS1[∆N∆C] lines (TPS1[∆N∆C] and TPS1[∆N∆C]-GFP) had smaller rosettes, much shorter roots, and severely delayed flowering, irrespective of whether this was scored as days to bolting or total leaf number (Figures 2 and 3; Supplemental Figures 4 to 6). Surprisingly, the TPS1[∆C] lines (TPS1[∆C] line no. 7, TPS1[∆C] line no. 8, and TPS1[∆C]-GFP), lacking only the C-terminal TPP-like domain, were even more severely compromised and closely resembled the TPS1[A119W] lines (TPS1[A119W] line no. 2 and TPS1[A119W] line no. 6). Given the similarity of the phenotypes conferred by TPS1[∆C] and TPS1[A119W], and their severity compared with the milder phenotype of the TPS1[∆N∆C] lines, we made an additional construct: TPS1[∆N A119W]. This construct was able to complement the tps1-1 mutant to yield viable seeds. Although dwarfed and slow-growing, the resulting plants were eventually able to flower (Figure 3). These observations showed that removal of the C-terminal TPP-like domain or substitution of Ala-119 by Trp has a particularly deleterious effect on the functionality of the TPS1 protein, but this is less severe if the N-terminal domain of the protein is also removed.
Given the severe phenotypes associated with the absence of the C-terminal domain of TPS1, we examined the TPS1 sequence in more detail to find possible clues as to its physiological function. We identified two predicted phosphorylation sites that have been experimentally confirmed in planta: Ser-826 [S822⋅R⋅P⋅S⋅(pS)⋅D⋅S⋅G⋅A⋅K831] and Ser-941 [L935⋅A⋅D⋅T⋅T⋅S⋅(pS)⋅P942], the latter being seen in two independent studies (Wang et al., 2013; Roitinger et al., 2015). The C-terminal domain also contains a putative sumoylation site on Lys-902 with a very high confidence score (P = 0.005) according to the SUMOSP prediction tool (Zhao et al., 2014; http://sumosp.biocuckoo.org/). Indeed, this site matches the consensus for sumoylation: Ψ⋅K⋅X⋅[D/E] (where Ψ is a hydrophobic residue and X = any residue). To assess the potential influence of the putative sumoylation (Lys-902) and Ser-941 phosphorylation sites, we generated an additional construct encoding a version of TPS1 with a 48-amino acid truncation of the distal part of the C-terminal domain (TPS1[ΔC895-942]). This construct was able to complement the tps1-1 mutant through embryogenesis, but the resulting TPS1[ΔC895-942] plants were severely dwarfed and did not flower (Figures 3D and 3E) and thus closely resembled the TPS1[ΔC] lines.
Tissue and Cellular Localization of Arabidopsis TPS1
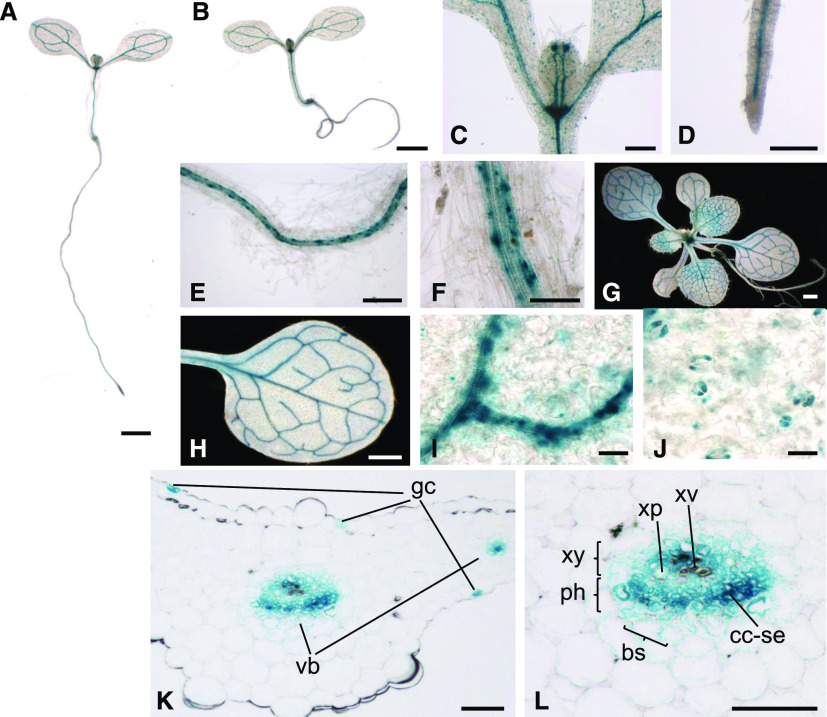
We grew seedlings and 2-week-old plants of the GUS-TPS1 and TPS1-GUS lines in long-day conditions and harvested them 10 h after dawn for detection of GUS activity. We observed the same expression patterns in both GUS-TPS1 (Figure 4A) and TPS1-GUS (Figure 4B) lines, and the results were consistent in multiple independent lines for each construct. Representative images from two independent N-terminal and two independent C-terminal GUS fusion lines are shown in Figure 4 and Supplemental Figure 7. The GUS-tagged TPS1 fusion protein was predominantly located in the leaf and root vasculature in seedlings and 2-week-old plants (Figures 4A to 4I) and in guard cells (Figure 4J). In transverse sections of the fourth true leaf from the GUS-TPS1 lines, the GUS-TPS1 fusion proteins were primarily located in the phloem tissue, especially in sieve elements and companion cells, with some weaker staining in the phloem parenchyma (bundle sheath cells) around the vascular bundle and in xylem parenchyma cells (Figures 4K to 4L). GUS-tagged TPS1 was also detected at the shoot apex (Figure 4C), consistent with previous RNA in situ hybridization data showing expression in the flanks of the SAM of vegetative plants (Wahl et al., 2013). In shoot apices from young, nonflowering plants, GFP-TPS1 localized to the peripheral and rib zones of the SAM (Figure 5C) and in the subtending vasculature (Supplemental Figure 8A). In inflorescence shoot apices, GFP-TPS1 was detected in floral primordia but not in the inflorescence meristem itself (Supplemental Figure 9). In the roots, TPS1 was found in companion cells and sieve elements of the phloem but not in the root apical meristem (Figures 4D to 4F).
Figure 4.
Tissue Localization Pattern of the Arabidopsis TPS1 Protein.
Full-length TPS1 fusion proteins tagged with GUS at either the N terminus (GUS-TPS1) or the C terminus (TPS1-GUS) were introduced into the tps1-1 mutant, grown in a 16-h photoperiod, and harvested 10 h after dawn for GUS activity staining.
(A) Seedling of the TPS1-GUS lines.
(B) Seedling of the GUS-TPS1 lines.
(C) to (J) Localization of GUS-tagged TPS1 in the shoot (C) and root ([D] to [F]) of seedlings and in the whole rosette (G) or fully expanded leaves ([H] to [J]) of 2-week-old soil-grown plants.
(K) and (L) Transverse sections of the fourth leaf showing GUS-tagged TPS1 in guard cells (gc) and vascular bundles (vb). bs, bundle sheath (phloem parenchyma); cc-se, companion cell/sieve element complex; ph, phloem; xp, xylem parenchyma; xv, xylem vessel (tracheid); xy, xylem.
Bars = 1 mm ([A], [B], [G], and [H]), 200 µm ([C], [D], and [E]), 50 µm ([F], [K], and [L]), and 20 µm ([I] and [J]).
Figure 5.
Subcellular Localization of Arabidopsis TPS1.
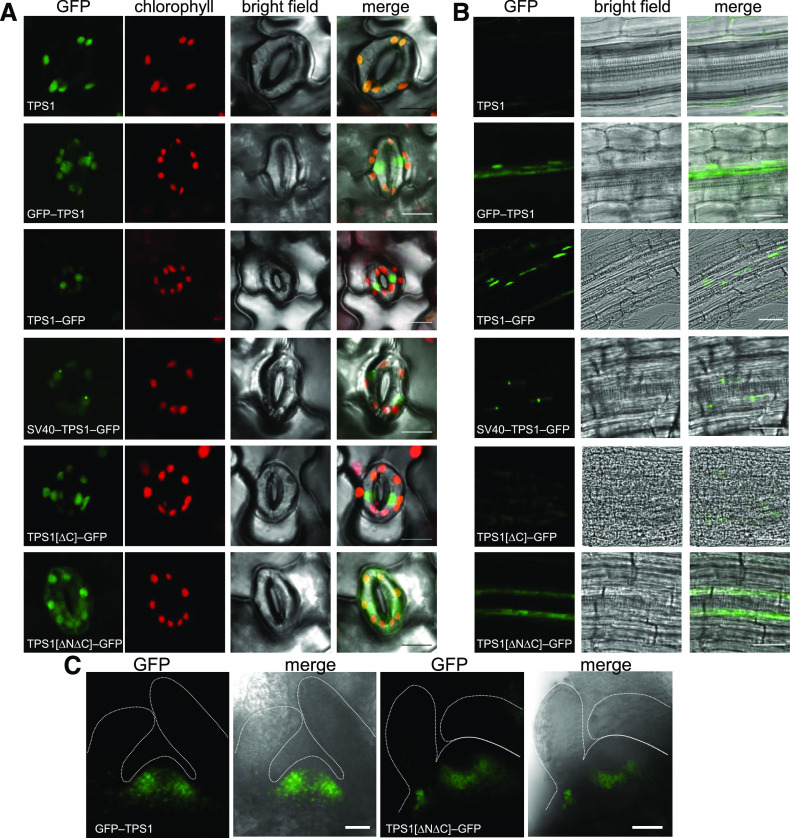
The tps1-1 mutant was complemented with various constructs encoding GFP-tagged forms of the TPS1 protein, and the localization of the GFP fusion proteins was examined by laser scanning confocal microscopy in guard cells on the abaxial surface of the leaf (A), roots of 8-d old seedlings (B), and SAMs (C). A complemented line expressing the native (i.e., nontagged) TPS1 protein (TPS1; top row) was used as a negative control for autofluorescence. The constructs used for complementation of tps1-1 are shown in Figure 1. The green and red channels show GFP and chlorophyll autofluorescence, respectively, and the merged images show the GFP signal superimposed on the bright-field images. In (C), the dotted white lines mark the outer surface of the meristem and leaf primordia. Bars = 30 μm.
We also looked in reproductive tissues of older GUS-TPS1 and TPS1-GUS plants. In unopened flower buds, we detected GUS-tagged TPS1 at the base of the flower, in the pedicel, and at the base of the receptacle (Supplemental Figures 7A to 7C). In open flowers, GUS staining was particularly prominent in the vasculature of petals, sepals, carpels, and stamens, in stamen guard cells, and in pollen, with each pollen grain having two distinct foci of GUS activity (Supplemental Figures 7D to 7J). In siliques, GUS-tagged TPS1 was present in the vasculature of the valves and in the abscission zone of the pedicel, as well as in the funiculus and the abscission zones of developing seeds (Supplemental Figures 7L to 7N). We detected no GUS activity in developing seeds prior to the globular stage of embryo development, but GUS-tagged TPS1 was detected in embryos from the mid to late globular stage (Supplemental Figure 7O), with the strongest expression in the middle zone of the globular embryo, which develops into the vascular tissue of the mature embryo. Tannin accumulation in the seed coat rendered the tissue impermeable to the 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid substrate, preventing us from detecting GUS-tagged TPS1 after the globular stage. We detected GUS activity in mature seeds (Supplemental Figure 7P), although it was only visible in the chalazal endosperm due to the tannin accumulation in the seed coat. However, after germination and rupture of the seed coat, GUS-tagged TPS1 was detected throughout the seed (Supplemental Figure 7Q), suggesting that TPS1 was probably already present in all of the seed tissues during the later stages of seed maturation. In mature Arabidopsis embryos, we observed a GUS signal in the radicle, the vasculature, and close to the SAM (Supplemental Figure 7R).
Subcellular Compartmentation of Arabidopsis TPS1
For microscopic analysis of the GFP-tagged TPS1 lines, we focused on tissues where a TPS1 signal would be strong and easily visible based on our GUS-tagged TPS1 lines (i.e., guard cells, seedling roots, and shoot apices). In guard cells, GFP strictly localized to the nuclei in both GFP-TPS1 and TPS1-GFP lines, with no apparent GFP signal outside the nucleus (Figure 5A). As the GFP signal was relatively weak compared with the autofluorescence of chlorophyll in the chloroplasts, we imaged the nontagged TPS1 line under the same conditions as a negative control (Figure 5A). We confirmed the nuclear localization of GFP-tagged TPS1 in guard cells by microscopic analysis of independently grown plants and staining of nuclei with 4′,6-diamidino-2-phenylindole (DAPI), which showed overlap of the GFP and DAPI fluorescence signals in the nucleus (Supplemental Figure 10). In seedling roots, we only observed GFP-TPS1 in the nuclei of phloem companion cells and, more diffusely, in sieve elements, consistent with a cytosolic location in the latter (Figure 5B). In both nonflowering and inflorescence shoot apices, the GFP signal had a punctate appearance, consistent with a predominantly nuclear localization (Figure 5C; Supplemental Figures 8 and 9).
The N-terminal domain of Arabidopsis TPS1 contains a seven-amino acid motif, L27⋅R⋅E⋅K⋅R⋅K⋅S33, that resembles the proposed consensus sequence for monopartite NLSs, K⋅(K/R)⋅X⋅(K/R) (Chelsky et al., 1989), and matches perfectly with a shorter K⋅R⋅K motif sufficient for nuclear localization (Kosugi et al., 2009). We also identified the K30⋅R⋅K32 motif of TPS1 as a putative NLS signal using the NLStradamus prediction tool (posterior threshold > 0.6; Nguyen Ba et al., 2009). In the TPS1[∆N∆C]-GFP line, we observed a diffuse GFP signal throughout the cytoplasm in guard cells, shoot apical cells, and root phloem companion cells (Figures 5A to 5C; Supplemental Figure 8B). By contrast, the TPS1[∆C]-GFP line, which retains the N-terminal domain, still showed nuclear localization of the C-terminally truncated TPS1 protein in guard cells (Figure 5A). We detected no GFP signal in roots of the TPS1[∆C]-GFP line, and the vascular bundles were disorganized in the roots of these plants (Figure 5B). These observations are consistent with the presence of an endogenous NLS in the N-terminal domain of TPS1.
The SV40-TPS1-GFP fusion protein in guard cells exclusively localized to the nuclei (Figure 5A; Supplemental Figure 10). In some cells, there was a single prominent focus in the GFP signal within the nucleus (Figure 5A), suggesting localization in the nucleolus. In the root phloem of SV40-TPS1-GFP lines, the GFP signal was likewise restricted to the nuclei of the companion cells, each with a single bright focus (Figure 5B). In contrast to the other lines, no GFP signal was detected in the sieve elements of the SV40-TPS1-GFP lines (Figure 5B), suggesting that the strong SV40-NLS trapped all of the SV40-TPS1-GFP protein expressed in the companion cell-sieve element complex in the nuclei of the companion cells.
Tre6P Levels and Metabolite Profiles of tps1-1 Complementation Lines Grown under Long-Day and Equinoctial Conditions
Tre6P exerts a powerful influence not only on plant growth and development but also on carbon and nitrogen metabolism (Figueroa et al., 2016). Therefore, we investigated whether the TPS constructs could restore wild-type-like metabolism to the plants during vegetative growth by measuring Tre6P and a wide range of other metabolites in the rosettes of plants grown in long-day conditions (16-h photoperiod). We harvested lines with wild-type or near-wild-type growth habits 17 d after sowing 10 h after dawn. The two very dwarfed lines, TPS1[∆C] and TPS1[A119W], were grown in a separate experiment, along with wild-type Col-0 and a TPS1-complemented line as controls, and also harvested 10 h after dawn (Zeitgeber time 10 [ZT10]) but on different days, such that the plants from all genotypes were sampled at the same developmental stage (seven to eight fully expanded leaves).
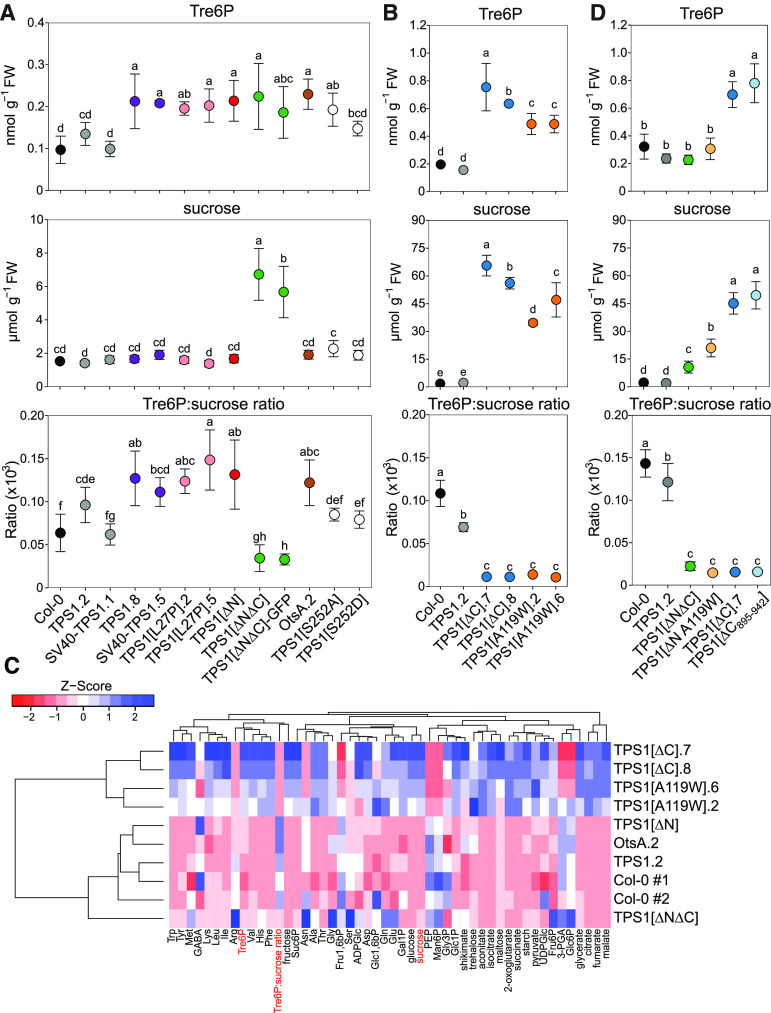
Tre6P levels in wild-type Col-0 rosettes ranged from 0.1 to 0.2 nmol g−1 fresh weight (Figures 6A and 6B), consistent with previous reports from plants grown under similar conditions (Lunn et al., 2006; Martins et al., 2013; Yadav et al., 2014; Figueroa et al., 2016). Tre6P levels were within the same range in all of the complementation lines except for TPS1[∆C] and TPS1[A119W] (Figures 6A to 6B). We had expected the A119W mutation to compromise the catalytic activity of the TPS1 protein, based on the failure of TPS1[A119W] to restore growth of the yeast tps1Δ mutant on glucose-containing media (Vandesteene et al., 2010). However, the TPS1[A119W] lines (0.50 nmol g−1 fresh weight) had more than twice as much Tre6P as Col-0 and TPS1 control plants (Figure 6B), demonstrating that the A119W mutation did not abolish catalytic activity. The TPS1[∆C] lines, which just like TPS[A119W] were dwarfed and nonflowering, also had elevated Tre6P levels (0.65 to 0.75 nmol g−1 fresh weight; Figure 6B).
Figure 6.
Analysis of Tre6P, Suc, and Other Metabolites in tps1-1 Complementation Lines Grown in Long-Day Conditions.
Wild-type Arabidopsis Col-0 plants and tps1-1 complementation lines were grown in a 16-h photoperiod. FW, fresh weight.
(A) Whole rosettes were harvested 10 h after dawn (ZT10) from plants 17 d after sowing for metabolite analysis.
(B) Two slow-growing lines, TPS1[ΔC] and TPS1[A119W], were grown in a separate experiment, along with wild-type Col-0 and a TPS1 line as controls, and whole rosettes were harvested 10 h after dawn (ZT10) from plants with seven or eight fully expanded leaves.
In (A) and (B), data for Tre6P, Suc, and the Tre6P:Suc ratio (×103) are shown as means ± sd (n = 5).
(C) Hierarchical clustering analysis of Tre6P, Suc, and other metabolite data (Supplemental Figures 11 and 12). Z-scores of the mean were calculated for each metabolite and are presented as a heatmap, with high and low Z-scores shown in blue and red. Col-0 no. 1 and Col-0 no. 2 represent the wild-type Col-0 plants from the experiments shown in (A) and (B). Dendrograms represent clusters of the lines or metabolites based on similarity using a distance matrix computed with the canberra distance measure.
(D) Two additional lines, TPS1[ΔN A119W] and TPS1[ΔC895-942], were grown in another experiment, along with wild-type Col-0, TPS1 (line no. 2), TPS1[ΔC] (line no. 7), and TPS1[ΔNΔC]. Whole rosettes were harvested 12 h after dawn from plants with six to eight fully expanded leaves. Data for Tre6P, Suc, and the Tre6P:Suc ratio (×103) are shown as means ± sd (n = 4).
In (A), (B), and (D), letters represent significant differences (P ≤ 0.05) based on ANOVA with posthoc LSD testing. Black symbols represent wild-type Col-0. Other symbol colors represent the tps1-1 mutant transformed with the constructs shown in Figure 1: TPS1 (gray), SV40-TPS1 (purple), TPS1[L27P] (pink), TPS1[ΔN] (red), TPS1[ΔNΔC] (±GFP) (green), OtsA (brown), TPS1[S252A] and TPS1[S252D] (white), TPS1[ΔC] (blue), TPS1[A119W] (orange), TPS1[ΔC895-942] (light blue), and TPS1[ΔN A119W] (light orange).
All complementation lines had wild-type levels of Suc (2 to 3 µmol g−1 fresh weight), except for the TPS1[∆N∆C] (±GFP) lines, which had about twice as much Suc as wild-type plants (6 µmol g−1 fresh weight), and the TPS1[A119W] (35 to 47 µmol g−1 fresh weight) and TPS1[∆C] (57 to 66 µmol g−1 fresh weight) lines (Figures 6A and 6B). The levels of Suc accumulated in these two lines are unprecedented, being up to 10 times higher than we have ever previously observed in Arabidopsis wild-type plants or sugar-accumulating mutants (Lunn et al., 2006; Dos Anjos et al., 2018). The Tre6P:Suc ratios in the TPS1[∆N∆C] (±GFP), TPS1[A119W], and TPS1[∆C] lines were up to 10-fold lower than in wild-type plants, driven mainly by their high Suc contents (Figures 6A and 6B). Sucrose 6′-phosphate (Suc6P), the intermediate of Suc biosynthesis, was significantly higher than the wild type in the TPS1[∆N∆C] (±GFP) lines (Supplemental Figure 11A) and even higher in the TPS1[∆C] and TPS1[A119W] lines (Supplemental Figure 11B), suggesting altered rates of Suc synthesis in these lines compared with wild-type plants.
Among the other metabolites measured, the TPS1[∆N] line had elevated levels of pyruvate and several tricarboxylic acid (TCA) cycle intermediates: citrate, aconitate, and 2-oxoglutarate (Supplemental Figure 11A). Aconitate and 2-oxoglutarate were also significantly increased in the OtsA line (Supplemental Figure 11A). We observed the most extreme differences from the wild type in the TPS1[A119W] and TPS1[∆C] lines. Both lines had elevated levels of most organic acids, with fumarate being increased up to 20-fold, while fructose-1,6-bisphosphate, mannose 6-phosphate, 3-phosphoglycerate, and phosphoenolpyruvate were significantly lower than in the controls (Supplemental Figure 11B).
We also analyzed individual amino acids in some of the lines. The TPS1[∆N∆C] line had elevated levels of Glu, Gln, Ala, Ser, Gly, Asn, Arg, Met, His, and Phe (Supplemental Figure 12). Almost all of the measured amino acids were also increased in the TPS1[∆C] lines and to a slightly lesser extent in the TPS1[A119W] lines (Supplemental Figure 12). By contrast, amino acid levels in the TPS1[∆N] line were not significantly different from the wild type, and only Gly was slightly increased in the OtsA line (Supplemental Figure 12).
To get an overview of the metabolic similarities and differences between the lines, we performed a hierarchical clustering analysis on the lines with the most comprehensive metabolite profiling and visualized the differences as a heatmap (Figure 6C). As might be expected, the dwarfed TPS1[∆C] and TPS1[A119W] lines diverged highly from the other lines, with most metabolites in the TPS1[∆C] and TPS1[A119W] lines showing opposite trends to those in the wild-type and other lines. Within the second cluster, which included all of the other lines analyzed, the TPS1[∆N∆C] line was a partial outlier, driven by its lower Tre6P:Suc ratio and differences in several amino acids and phosphorylated intermediates. The TPS1[∆N] and OtsA lines were the most similar to the wild-type Col-0 and TPS1 controls.
In a separate experiment, we grew the TPS1[ΔNΔC], TPS1[ΔN A119W], TPS1[ΔC], and TPS1[ΔC895-942] lines, along with wild-type (Col-0) and wild-type-like (TPS1 line no. 2) plants, in long-day (16-h photoperiod) conditions and harvested rosettes 12 h after dawn for metabolite analysis. There were no significant differences between wild-type and TPS1 line no. 2 plants in their Tre6P and Suc contents (Figure 6D), and these were similar to the respective values seen in previous experiments (Figures 6A and 6B). The TPS1[ΔNΔC] line had moderately elevated Suc levels and a lower Tre6P:Suc ratio than wild-type plants (Figure 6D), as previously observed (Figure 6A). Similarly, the TPS1[ΔN A119W] lines had moderately high Suc and a lower Tre6P:Suc ratio than wild-type plants (Figure 6D). In contrast, TPS1[ΔC] line no. 7 had exceptionally high Tre6P and Suc levels and a low Tre6P:Suc ratio (Figure 6D), confirming previous results (Figure 6B). We saw a very similar pattern in the TPS1[ΔC895-942] plants (Figure 6D). We confirmed the strong similarity between the TPS1[ΔC895-942] and TPS1[ΔC] no. 7 lines, both of which have severely dwarfed and nonflowering phenotypes, by performing a broader metabolite analysis, which showed these two lines clustering closely together and being well separated from the wild-type plants and the two lines with less-severe growth phenotypes (TPS1[ΔNΔC] and TPS1[ΔN A119W]; Supplemental Figure 13).
We also grew some of the lines under equinoctial conditions (12-h photoperiod) to see if further metabolic differences emerged when the plants are more carbon-limited than in long-day conditions. We harvested rosettes from 4-week-old plants at the end of the day and at the end of the night for metabolite analysis (Supplemental Figure 14). As seen already in long-day conditions, the dwarfed TPS1[∆C] and TPS1[A119W] lines were the most divergent. Their Tre6P levels were mostly not significantly different from the wild type or only moderately increased, but they accumulated up to 30 times more Suc than wild-type Col-0 and TPS1 control plants and therefore had lower Tre6P:Suc ratios (Supplemental Figure 14B). The two dwarfed lines also had more starch at the end of the day and a starch excess at the end of the night (Supplemental Figure 15A). In comparison, the metabolite profiles of all the other analyzed lines were similar to wild-type Col-0 and to each other. Tre6P was moderately lower in the TPS1[∆N∆C] lines at the end of the day and approximately twofold higher in TPS1[L27P] line no. 5 at the end of the night (Supplemental Figure 14A). TPS1[∆N∆C] (±GFP) lines had twice as much Suc as the other lines at both times of day, leading to a lower Tre6P:Suc ratio, and the OtsA line showed a similar trend (Supplemental Figure 14A). Hierarchical clustering and heatmap analysis confirmed the divergence of TPS1[∆C] and TPS1[A119W] from all other lines, with most metabolites in these two lines showing opposite behavior compared with all other genotypes (Supplemental Figures 14C and 14D).
Relationship between Tre6P and Suc in tps1-1 Complementation Lines
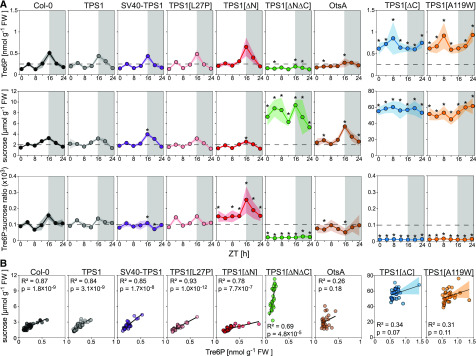
One of the most intriguing questions was whether the relationship between Tre6P and Suc was preserved in the tps1-1 complementation lines. To address this question, we grew representative lines in long-day conditions and harvested 20-d-old plants at 4-h intervals over a complete 24-h light-dark cycle for metabolite analysis. Both wild-type Col-0 and TPS1 plants showed very similar diurnal patterns, with Tre6P and Suc increasing in parallel over the day and falling at night (Figure 7A). The Tre6P:Suc ratio fluctuated within a relatively narrow range (Figure 7A), and there was a strong and highly significant correlation between Tre6P and Suc in both Col-0 (R2 = 0.87, P = 1.8 × 10−9) and TPS1 (R2 = 0.84, P = 3.1 × 10−9; Figure 7B). The SV40-TPS1 and TPS1[L27P] lines were very similar to the Col-0 and TPS1 plants. The TPS1[∆N] lines had a slightly higher Tre6P:Suc ratio (Figure 7A). Tre6P was still correlated with Suc (R2 = 0.78, P = 7.7 × 10−7), although the slope of the regression line was shallower (Figure 7B), resembling the pattern seen in plants with constitutive overexpression of TPS (Yadav et al., 2014). The TPS1[∆N∆C] line had 3 to 5 times more Suc than Col-0, while Tre6P and the Tre6P:Suc ratio were generally lower than in Col-0. Tre6P was still fairly well correlated with Suc (R2 = 0.69, P = 4.8 × 10−5), but the slope of the regression line was much steeper, resembling the pattern seen in plants with constitutive overexpression of TPP (Yadav et al., 2014). Although Tre6P levels in the OtsA line were in the same range as Col-0 plants, the diurnal fluctuations in Tre6P appeared to be dampened in the OtsA line, with Tre6P being slightly higher than in wild-type plants in the middle of the day (Figures 6A and 7A) but lower at the end of the day (Figure 7A; Supplemental Figure 16A). By contrast, their Suc content showed a greater diurnal amplitude, with higher levels than in wild-type and TPS1-complemented plants at the end of day (Figure 7A; Supplemental Figure 16A). There was no significant correlation between Tre6P and Suc in the OtsA plants (R2 = 0.26, P = 0.18; Figure 7B). The dwarfed TPS1[∆C] and TPS1[A119W] lines again showed the greatest differences from the wild-type Col-0 controls and other lines. They had 3 to 5 times more Tre6P and up to 30 times more Suc than Col-0 plants throughout the diurnal cycle, giving them a very low Tre6P:Suc ratio (Figure 7A). There was no significant correlation between Tre6P and Suc in these two lines (Figure 7B).
Figure 7.
Diurnal Profiles and Correlation of Tre6P and Suc in tps1-1 Complementation Lines Grown in Long-Day Conditions.
Wild-type Arabidopsis Col-0 plants and tps1-1 complementation lines were grown in a 16-h photoperiod. FW, fresh weight.
(A) Whole rosettes were harvested at 4-h intervals starting at dawn (ZT0) from plants at 20 d after sowing for metabolite analysis. Data for Tre6P, Suc, and the Tre6P:Suc ratio (×103) are presented as means ± sd (n = 4 biological replicates at each time point). Asterisks indicate significant differences (P ≤ 0.05) from the wild-type Col-0 and a TPS1-complemented line (TPS1 line no. 2) based on ANOVA with posthoc LSD testing. Data for other metabolites from the same experiment are shown in Supplemental Figures 15B and 16.
(B) Pearson correlation analysis of Tre6P and Suc data (note that all individual samples from the experiment shown in [A] are included [i.e., four biological replicates per time point]). Black symbols represent wild-type Col-0. Other symbol colors represent the tps1-1 mutant transformed with the constructs shown in Figure 1: TPS1 (line no. 2; gray), SV40-TPS1 (line no. 1; purple), TPS1[L27P] (line no. 5; pink), TPS1[ΔN] (red), TPS1[ΔNΔC] (green), OtsA (line no. 2; brown), TPS1[∆C] (line no. 7; blue), and TPS1[A119W] (line no. 6; orange).
The diurnal profiles of other sugars, phosphorylated intermediates, organic acids, and starch were essentially identical in the Col-0 and TPS1 plants (Supplemental Figures 15B and 16) and very similar in the SV40-TPS1 and TPS1[L27P] lines (Supplemental Figures 16A and 16B). The TPS1[∆N] line had more trehalose and slightly elevated levels of TCA cycle intermediates, especially at night, but was otherwise similar to Col-0 (Supplemental Figure 16C). The TPS1[∆N∆C] line showed greater differences, with generally higher levels of phosphorylated intermediates and lower TCA cycle intermediates (Supplemental Figure 16D). The OtsA line had higher Suc6P at night and lower fumarate during the day but was otherwise very similar to Col-0 (Supplemental Figure 16E). In the dwarfed TPS1[ΔC] and TPS1[A119W] lines, almost all metabolites, including starch (Supplemental Figure 15B), were significantly higher than in Col-0 throughout the diurnal cycle, except for pyruvate, which was 2 to 5 times lower in the two complementation lines compared with Col-0 (Supplemental Figures 16F and 16G).
Response of Tre6P to Suc in Carbon-Starved Seedlings
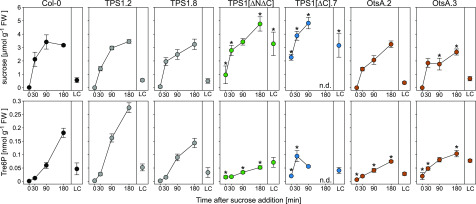
In three of the tps1-1 complementation lines—OtsA, TPS1[∆C], and TPS1[A119W]—we observed that Tre6P did not follow the endogenous diurnal fluctuations in Suc levels (Figure 7), suggesting that the sucrose-Tre6P nexus relationship was broken in these lines. To investigate this further, we tested whether Tre6P levels changed when Suc was supplied exogenously to carbon-starved seedlings. As observed in previous studies (Lunn et al., 2006; Yadav et al., 2014), Tre6P was very low in carbon-starved Col-0 seedlings but, after a short lag, increased rapidly after Suc addition (Figure 8). Two independent TPS1-complemented lines, TPS1 line no. 2 and TPS1 line no. 8, showed a behavior similar to wild-type Col-0 seedlings. The changes in sugar levels in two OtsA lines, OtsA line no. 2 and OtsA line no. 3, were broadly similar to those of Col-0 seedlings (Figure 8; Supplemental Figure 17A), but there was a weaker response of Tre6P to Suc addition (Figure 8). Both sets of lines containing a deletion of the C-terminal domain—TPS1[∆N∆C] and TPS1[∆C]—accumulated higher levels of Suc than Col-0 (Figure 8), but the increase in Tre6P levels in these lines was much weaker and less consistent than in the Col-0 and TPS1 seedlings. For the other metabolites that we measured, the responses to Suc addition were similar to Col-0 in all lines except for TPS1[∆C] (Supplemental Figure 17B), as it showed a greater increase in phosphoenolpyruvate after Suc was supplied but smaller increases in most TCA cycle intermediates: citrate, aconitate, isocitrate, 2-oxoglutarate, succinate, and malate.
Figure 8.
Response of tps1-1 Complementation Seedlings to Exogenous Suc Supply.
Wild-type Col-0 plants and tps1-1 complementation lines were grown on solid half-strength Murashige and Skoog medium (without Suc) in constant light (150 μmol m−2 s−1 irradiance) at 22°C for 10 d, dark-adapted for 2 d to induce carbon starvation, and then transferred to liquid half-strength Murashige and Skoog medium containing 15 mM Suc. Whole seedlings were harvested at 0, 30, 90, and 180 min after transfer to Suc-containing medium for metabolite analysis. LC, light control (10-d-old nonstarved seedlings). Suc and Tre6P data are presented as means ± sd (n = 3 or 4 biological replicates at each time point). Asterisks indicate significant differences (P ≤ 0.05) from the three control genotypes, wild-type Col-0 and two independent TPS1-complemented tps1-1 lines (TPS1 line no. 2 and TPS1 line no. 8), based on ANOVA with posthoc LSD testing. Other metabolite data from the same experiment are shown in Supplemental Figure 17. Black symbols represent wild-type Col-0. Other symbol colors represent the tps1-1 mutant transformed with the constructs shown in Figure 1: TPS1 (gray), TPS1[ΔNΔC] (green), TPS1[∆C] (blue), and OtsA (brown). FW, fresh weight; n.d., not determined.
The Dwarfed TPS1[ΔC] and TPS1[A119W] Lines Accumulate Novel Disaccharide-Monophosphates
To measure Tre6P, we used an anion-exchange HPLC coupled to tandem mass spectrometry (LC-MS/MS)-based assay that gives a baseline resolution of Tre6P and its most common isomer in plants, Suc6P, the intermediate of Suc biosynthesis (Lunn et al., 2006). In addition to Tre6P and Suc6P, we have also observed several other compounds with the mass spectral properties of disaccharide-monophosphates in plant extracts. These include abundant levels of maltose 1-phosphate in orange (Citrus sinensis) leaves infected with a bacterial pathogen (Xanthomonas citri subsp citri; Piazza et al., 2015) and trace amounts of three other isomeric molecules in Arabidopsis plants (Supplemental Figure 18), the identities of which are so far unknown (discussed in Figueroa and Lunn, 2016). We noted that the levels of two of these unknowns, which elute just before and just after Suc6P (Supplemental Figure 18A), were massively elevated in the dwarfed TPS1[∆C] and TPS1[A119W] lines in all experiments and also slightly higher than the wild type in the TPS1[∆N∆C] (±GFP) lines (Supplemental Figure 18). As their identities are currently unknown, we had no standards for calibration, so we calculated their concentrations based on the calibration curve for Suc6P, which had the most similar retention time. We estimated that, compared with wild-type and TPS1 control plants, the levels of these two compounds were 3 to 4 times higher in the TPS1[∆N∆C] (±GFP) lines, 10 to 20 times higher in TPS1[∆N A119W], and up to 50 times higher in the TPS1[∆C], TPS1[∆C895-942], and TPS1[A119W] lines (Supplemental Figure 18). In common with Tre6P, these compounds strongly increased after Suc addition to carbon-starved TPS1[∆C] seedlings (Supplemental Figure 18E).
DISCUSSION
Our study aimed to answer five questions. (1) Why is TPS1 essential for embryogenesis in Arabidopsis? (2) In which tissues and cell types is TPS1 located? (3) Where in the cell is TPS1 protein localized? (4) What are the functions of the noncatalytic domains of the TPS1 protein? (5) How does TPS1 contribute to the sucrose-Tre6P nexus?
The Essential Role of TPS1 in Embryogenesis
The defective embryogenesis in Arabidopsis tps1 null mutants (Eastmond et al., 2002) was one of the first discoveries that demonstrated the importance of the trehalose biosynthetic pathway for plant growth and development. As a result, what had once been just an obscure branch of sugar metabolism, of little interest except to dedicated sugar biochemists, began to receive much greater attention. Subsequent discoveries revealed how ubiquitous the influence of trehalose metabolism is; perturbation of this pathway leads to alterations in carbon and nitrogen metabolism, leaf development, flowering time, shoot branching, stomatal function, and tolerance of abiotic and biotic stresses, not only in Arabidopsis but also in other plant species (reviewed in Figueroa and Lunn, 2016). The primary defect in tps1-1 null mutants appeared to be a lack of Tre6P, based on reports that the tps1-1 mutant could be rescued by expression of the heterologous E. coli otsA gene under the control of the presumed Arabidopsis TPS1 promoter (Schluepmann et al., 2003; van Dijken et al., 2004). However, the complementation construct used in those studies to drive otsA expression was based on an incomplete annotation of the TPS1 locus, such that the presumed TPS1 promoter was in reality part of the first intron and 5′-UTR (as indicated in Figure 1A). It is therefore surprising that the otsA gene was expressed at all in this construct, but one possible explanation is that otsA expression was driven by read-through transcription from the constitutive promoter of the selectable marker gene in the T-DNA. Whatever the mechanism driving otsA expression in those previous studies, its spatiotemporal expression pattern would have differed from that of the endogenous TPS1 gene. Furthermore, there was minimal documentation of the phenotypes of the otsA complementation lines in the published reports, leaving many questions unanswered. Previous examination of arrested tps1 embryos revealed defects in cell division, cell wall biosynthesis, and accumulation of storage reserves (Eastmond et al., 2002; Gómez et al., 2006), but a fundamental question remained: is the principal defect in the tps1 mutants a lack of Tre6P synthesis in specific cells at a crucial stage of embryo development, or is it linked to some noncatalytic function of TPS1, or both?
The full complementation of the tps1-1 mutant by the TPS1pro:otsA:TPS1term construct provided the answer. This demonstrated that restoring Tre6P synthesis in the cell types and developmental stages where TPS1 is expressed during embryogenesis was sufficient to rescue the tps1-1 mutant through seed development to yield viable seeds. With the exception of TPS1[R369A,K374A,E476A], all of the constructs encoding wild-type, tagged, mutated, or truncated forms of TPS1 also rescued the tps1-1 mutant through embryogenesis. This included the TPS1[A119W] construct encoding a form of TPS1 that, unlike wild-type TPS1 (Blázquez et al., 1998), is unable to complement the yeast tps1Δ mutant by restoring Tre6P synthesis (Van Dijck et al., 2002). The failure of TPS1[A119W] to complement the yeast tps1Δ mutant suggested that this single-point mutation compromises the catalytic activity of TPS1. However, TPS1[A119W] was able to rescue the Arabidopsis tps1-1 mutant through embryogenesis, and the resulting plants contained measurable amounts of Tre6P that reached levels higher than wild-type plants (Figures 6 and 7). This showed that the A119W mutation does not abolish the catalytic activity of TPS1, even though it severely compromises TPS1 protein functionality in planta and, by inference, when expressed heterologously in yeast. All other mutated or truncated forms of TPS1 that were able to complement the tps1-1 mutant also retained catalytic activity, as the respective complementation lines had either normal or elevated Tre6P levels (Figures 6 and 7). The dependence of tps1-1 complementation on restoration of Tre6P synthesis was corroborated by the failure of the TPS1[R369A,K374A,E476A] construct, encoding a catalytically inactive TPS1, to rescue the tps1-1 mutant through embryogenesis. We conclude that the capacity to synthesize Tre6P during embryogenesis is both necessary and sufficient to rescue the tps1-1 mutant during seed development.
Tre6P appears to be dispensable in developing seeds until the torpedo stage, when tps1-1 embryos arrest (Eastmond et al., 2002). The torpedo stage of embryo development coincides with cellularization of the peripheral endosperm surrounding it, a process that is regulated by auxin (Batista et al., 2019). A major function of the cellularized endosperm is to nourish the embryo during later stages of development. Based on microarray data, the TPS1 gene is expressed in the embryo throughout seed development and is strongly upregulated in the peripheral endosperm at the torpedo stage (Schmid et al., 2005; Figueroa and Lunn, 2016). We might speculate that a lack of Tre6P in the embryo and/or peripheral endosperm at the torpedo stage disrupts cellularization of the endosperm, perhaps by perturbing auxin synthesis (Meitzel et al., 2019) or signaling, and thereby prevents the endosperm from fulfilling its function to nourish the developing embryo, leading to eventual embryo arrest. Other functional class I genes, TPS2 and TPS4, are expressed primarily in the chalazal endosperm (Schmid et al., 2005; Figueroa and Lunn, 2016). Thus, we conclude that, despite being catalytically active and expressed in developing seeds, the inability of the TPS2 and TPS4 isoforms to compensate for the loss of TPS1 in tps1 mutants is simply due to their lack of expression in the right cells at the right time to substitute for TPS1.
Tissue and Cellular Localization of TPS1
From transcriptomic analysis, TPS1 mRNA is expressed in all major organs of the plant: leaves, roots, flowers, and seeds (Supplemental Figure 1; Schmid et al., 2005). However, the transcript data had limited resolution at the tissue and cellular levels and do not necessarily reflect the distribution of the functional TPS1 protein if it is subject to posttranscriptional regulation. Our GUS/GFP-tagged TPS1 lines not only reported the localization of TPS1 itself but also provided greater tissue and cellular resolution. Both N- and C-terminally tagged versions of TPS1 fully complemented the tps1-1 mutant (Supplemental Figures 2 and 4) and showed the same expression patterns (Figures 4 and 5; Supplemental Figure 7), indicating that the tagged TPS1 proteins were fully functional and reliable reporters of the cellular and subcellular localization of TPS1.
In leaves, TPS1 predominantly accumulated in and around the vascular bundles and in guard cells (Figures 4 and 5). The latter confirms the detection of TPS1 in the guard cell proteome (Zhao et al., 2008) and corroborates the defective stomatal phenotypes of the tps1-12 mutant, which carries a weak but non-embryo-lethal tps1 allele (Gómez et al., 2010). The functions of TPS1 and Tre6P in guard cells are not yet understood; they appear to have an essential, but so far undefined, role in abscisic acid signaling (Gómez et al., 2010) and are also implicated in the regulation of various aspects of guard cell carbon metabolism (reviewed in Figueroa and Lunn, 2016; Santelia and Lunn, 2017).
In the leaf vasculature, TPS1 was seen in the phloem parenchyma (bundle sheath) cells around the vascular bundles as well as in the companion cells and sieve elements of the phloem itself (Figures 4K and 4L). One of the main functions of the phloem is to translocate Suc from source leaves to sink organs. Suc is produced by photosynthesis in the mesophyll cells during the day and by mobilization of transitory starch reserves at night. In Arabidopsis, Suc is actively loaded from the apoplast into the phloem for export to growing sink organs (Figure 9A; Haritatos et al., 2000). Suc diffuses via plasmodesmata from mesophyll cells into phloem parenchyma cells surrounding vascular bundles, where it is released into the apoplast by two SUCROSE WILL EVENTUALLY BE EXPORTED (SWEET)-type sugar transporters: SWEET11 and SWEET12 (Chen et al., 2012). Suc is then actively taken up from the apoplast into companion cells and sieve elements by SUT(SUC)-type sucrose-H+ symporters. Thus, TPS1 is located on both sides of the apoplastic barrier in the phloem-loading zone in the leaves, which essentially represents the interface between source and sink tissues, and is potentially a highly strategic site for signaling between source and sink (Figure 9A). For example, if the supply of Suc from the leaves exceeds the demand from growing sink organs, Suc will accumulate in the leaves. Rising Suc levels in leaf phloem parenchyma cells will lead to an increase in Tre6P, which can diffuse symplastically, via plasmodesmata, into mesophyll cells (Figure 9A) to shift the partitioning of photoassimilates away from Suc (Figueroa et al., 2016) or inhibit transitory starch turnover (Martins et al., 2013; Dos Anjos et al., 2018), thereby restoring the balance between Suc supply and demand.
Figure 9.
Localization of TPS1 in the Phloem-Loading Zone and Potential Regulation of TPS1 Activity in Arabidopsis.
(A) Schematic diagram of TPS1 expression in the phloem parenchyma (bundle sheath) and companion cell/sieve element complex in leaf vascular bundles and in the SAM. Tre6P synthesized in the phloem parenchyma can diffuse symplastically into mesophyll cells to regulate photoassimilate partitioning (day) and transitory starch turnover (night) according to the demand for Suc. Tre6P synthesized in the companion cells can diffuse symplastically into the sieve elements and be transported around the plant, potentially acting as a systemic signal of Suc availability from source leaves.
(B) TPS1 has three domains: an N-terminal domain (red), a catalytic glucosyltransferase (TPS) domain (gray), and a TPP-like domain (blue). Important amino acid residues for the NLS, phosphorylation sites (P), and the sumoylation site (S) identified in this study are indicated. Colored bars below TPS1 indicate predicted disordered regions (black) and regions that are homologous with bacterial TPS (OtsA; dark gray) and TPP (OtsB; dark blue) enzymes.
(C) Schematic representation of TPS1 structure, based on the known structures of bacterial OtsA (Gibson et al., 2002, 2004) and OtsB (Rao et al., 2006) proteins, showing potential mechanisms of regulation by Suc and Tre6P. Left, potential allosteric activation of TPS1 by Suc binding to the C-terminal domain, possibly involving interaction with a putative activator protein (Protein X; Yadav et al., 2014). Right (top), competitive inhibition by Tre6P binding in the active site of TPS1. Right (bottom), potential allosteric inhibition of TPS1 by Tre6P binding to the C-terminal domain, conceivably involving interaction with a hypothetical inhibitory protein (Protein Y). These scenarios are not mutually exclusive and would allow a tight control of TPS1 activity based on the sucrose:Tre6P ratio in a given cell. The intracellular localization of TPS1 might also affect its activity and is potentially regulated by modification or interactions of other proteins with the NLS in the N-terminal domain.
In companion cells, TPS1 is ideally situated to monitor the uptake and availability of Suc for export to sink organs and modulate Tre6P levels in the phloem accordingly. FLOWERING LOCUS T (FT) is a florigenic, phloem-mobile protein that is produced in companion cells under inductive long-day conditions (Chen et al., 2018) and moves to the SAM, where it triggers the transition to flowering. FT expression depends on functional TPS1 (Wahl et al., 2013). Our observation that the otsA-complemented tps1-1 plants flower at the same chronological age and leaf number as wild-type plants (Figure 2B), in the absence of any Arabidopsis TPS1 protein, shows that the dependence of FT expression on TPS1 is solely due to its Tre6P-synthesizing capacity. TPS1 in companion cells has the potential to influence other systemic signals that originate there. Tre6P itself is potentially a systemic signal, as it is almost certainly present in phloem sieve elements, given their symplastic connection with companion cells and the presence of TPS1 protein in sieve elements (Figures 4K and 4L), and is likely to be carried from source to sink tissues by the mass flow of solutes in the phloem (Figure 9A). Whether it has a signaling function when it arrives in sink organs remains an open question, which we are currently addressing in heterografting experiments.
TPS1 expression was seen in a range of sink tissues: (1) in the peripheral and rib zones of the vegetative SAM (Figure 5C); (2) in (proto)vasculature subtending the meristem (Supplemental Figure 8); (3) in root phloem (Figure 5B); (4) in floral primordia in the inflorescence SAM (Supplemental Figure 9); (5) in the vasculature and guard cells of floral tissues and siliques (Supplemental Figure 7); (6) in pollen (Supplemental Figure 7K); (7) in developing embryos (Supplemental Figure 7O); and (8) in germinating seeds, especially in the SAM and radicle (Supplemental Figure 7R). The functions of TPS1 and Tre6P in the SAM are poorly understood. Under short-day conditions, they act in the vegetative SAM to trigger flowering via interactions with the age-dependent pathway, mediated by the microRNA miR156 and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE proteins (SPL3 to SPL5; Wahl et al., 2013). In peripheral and rib zones of the SAM where TPS1 is present, cells undergo division, differentiation, and expansion, so TPS1 and Tre6P are potentially implicated in any, or all, of these processes. The altered cell number of tps1 mutant embryos (Gómez et al., 2006) and a report that TPS1 forms a complex with cell cycle-associated proteins—cyclin-dependent kinase A;1 (CDKA;1) and KCA1, a CDKA;1-interacting kinesin (Geelen et al., 2007)—appears to argue for a role for the TPS1 in cell division.
The presence of TPS1 in (proto)vasculature subtending the SAM suggests a role for TPS1 in vascular development. This is supported by the disrupted vascular bundles in roots of TPS1[∆C]-GFP lines (Figure 5B), which show no fluorescence signal for the tagged protein, indicating that a functional TPS1 is needed for proper development of root vasculature. In siliques, TPS1 accumulation was prominent in phloem-unloading tissues where Suc is delivered to developing seeds (Supplemental Figures 7D, 7E, and 8). These locations in growing sink organs are all places where cells are either undergoing differentiation and expansion or accumulating storage reserves (e.g., starch and oil), all of which are carbon- and energy-demanding processes that are dependent on Suc supplies. The presence of TPS1 in these locations suggests that Tre6P is synthesized in situ, signaling the local availability of Suc and regulating its utilization for growth and storage product synthesis (Schluepmann et al., 2003; Zhang et al., 2009; Zhai et al., 2018). We speculate that leaf-derived Tre6P arriving via the phloem might supplement Tre6P produced locally to reinforce the signaling of abundant Suc supplies, thereby integrating putative systemic and local Tre6P signals.
Subcellular Localization of TPS1
Wild-type TPS1 tagged with GFP (GFP-TPS1 and TPS1-GFP) localized predominantly in the nucleus of guard cells and companion cells and in the cytosol of phloem sieve elements (Figures 5A and 5B; Supplemental Figure 10). GFP-tagged TPS1 also had a punctate distribution in SAM cells (Figure 5C; Supplemental Figure 9), consistent with a predominantly nuclear localization in these cells as well. Mature pollen grains from the GUS-tagged TPS1 lines showed GUS activity in two subcellular bodies within the grains (Supplemental Figure 7K), consistent with TPS1 protein localization in the nuclei of both vegetative and generative cells. Due to the autofluorescence of chloroplasts in guard cells, we could not exclude the possibility that a small fraction of GFP-tagged TPS1 might not be found in chloroplasts, but other considerations suggest that TPS1 is not targeted to the chloroplasts. First, sequence-based prediction tools find no evidence of a chloroplast transit peptide at the N terminus of TPS1, and only full-length (106-kD) TPS1 was detected in immunoblots of wild-type Col-0 leaf extracts (Supplemental Figure 3), so there is no evidence of import into chloroplasts and subsequent signal peptide removal. Second, the observed nuclear localization of TPS1 in guard cells and phloem companion cells is consistent with the presence of a monopartite NLS (K30⋅R⋅K32) in the N-terminal domain of the protein that matches the minimal consensus sequence for such localization signals (Kosugi et al., 2009). Comparison of TPS1[∆N∆C]-GFP and TPS1[∆C]-GFP lines showed that removal of the N-terminal domain, containing the putative NLS, abolished targeting of the protein to the nucleus (Figures 5A and 5B; Supplemental Figure 10). We conclude that TPS1 primarily localizes to the nucleus of most cell types where it occurs and is targeted to the nucleus by a monopartite NLS in the N-terminal domain.
Why TPS1 localizes to the nucleus remains to be elucidated. The pores in the nuclear membrane are large enough to allow free movement of UDP-glucose, Glc6P, Tre6P, and UDP between the nucleus and cytosol, so there is no obstacle to Tre6P being synthesized in the nucleus and subsequently moving to the cytosol, or vice versa. It is well documented that changes in Suc content affect transcript levels of thousands of genes (Bläsing et al., 2005; Osuna et al., 2007; Cookson et al., 2016). As a proxy for Suc status, Tre6P is potentially involved in transcriptional regulation of gene expression by Suc, and the synthesis of Tre6P directly in the nucleus might allow more precise control over gene expression. The possibility that TPS1 itself binds to and modulates transcriptional regulators cannot be excluded. A further possibility is that TPS1 activity is regulated by association with other proteins that are located in the nucleus or the cytosol (Figure 9C). The failure of OtsA to fully complement the tps1-1 mutant suggests that such protein-protein interactions are not essential during embryogenesis but might be more important at later stages in development.
Phloem sieve elements are usually enucleate and have few ribosomes, and thus they are dependent on companion cells for mRNA and protein synthesis. In vascular bundles of the SV40-TPS1-GFP line, GFP signal was observed only in the nuclei of phloem companion cells, with no detectable signal in the sieve elements (Figure 5B). This indicated that the heterologous (SV40) NLS targeted TPS1 more strongly to the nucleus than the putative endogenous NLS (K30⋅R⋅K32) on its own, essentially trapping SV40-TPS1-GFP protein in the companion cell nuclei. Presumably, SV40-TPS1-GFP mRNA produced in companion cells would still be able to move into sieve elements, but if SV40-TPS1-GFP transcripts were translated by the few ribosomes present in the sieve elements, this would yield too little SV40-TPS1-GFP protein to be detected. Thus, we conclude that TPS1, when made in companion cells, is primarily targeted to the nucleus. We speculate that the endogenous monopartite NLS in TPS1 is not strong enough to target all of the protein to the nuclei of companion cell, potentially allowing some of the TPS1 protein to move symplastically into sieve elements via plasmodesmata. The SV40-TPS1-GFP line had shorter roots than wild-type Col-0 plants (Supplemental Figure 5) but, unlike the TPS1[∆C]-GFP line, the structure of the vascular bundles in the SV40-TPS1-GFP line was not obviously disrupted (Figure 5B). Therefore, we conclude that cytosolic TPS1 in sieve elements does have a purpose, as loss of TPS1 in these cells impairs root growth, and that this is linked to the functioning of differentiated phloem rather than the development of the vasculature in roots.
Functions of the Noncatalytic Domains of TPS1
The three-domain structure of TPS1 (Figure 1B) has ancient roots; class I TPS proteins in chlorophyte green algae also have the same three domains, as do orthologous proteins from bryophytes, lycophytes, ferns, and gymnosperms (Lunn, 2007). The retention of this three-domain structure throughout plant evolution indicates that it might have some purpose. Curiously, the analogous enzyme of Suc biosynthesis, sucrose-phosphate synthase (EC 2.4.1.14), has a C-terminal domain that resembles the next enzyme in that pathway, sucrose-phosphate phosphatase (SPP; EC 3.1.3.24), but has no catalytic activity (Lunn et al., 2000), reminiscent of the TPP-like C-terminal domain of TPS1.
In addition to its putative monopartite NLS described above (Figure 9B), the N-terminal domain of TPS1 has been proposed to have an autoinhibitory function, based on complementation assays in yeast (Van Dijck et al., 2002). This putative function is (1) linked to a Leu/Arg-rich motif (R20⋅L⋅R⋅D⋅R⋅E⋅L⋅R28) and (2) disrupted by substitution of Leu-27 by Pro (Van Dijck et al., 2002). We observed slightly elevated Tre6P in the TPS1[L27P] line in two experiments (Figure 6; Supplemental Figure 12), but in a more detailed diurnal time-course experiment, the plants had wild-type levels of Tre6P and trehalose throughout the light-dark cycle (Figure 7A; Supplemental Figure 16B), suggesting no major increase in flux through the pathway. The TPS1[L27P] plants had no obvious developmental phenotypes (Figure 2; Supplemental Figures 4 and 5) or differences in other metabolites (Supplemental Figures 11A, 12, 14, and 16B). From this, we conclude that the putative autoinhibitory region in the N-terminal domain has little influence on the catalytic activity of TPS1 in planta and no impact on plant growth and development, at least under our experimental growth conditions. Nevertheless, TPS1[∆N] plants showed a stronger phenotype, and this might be partly due to complete removal of the putative autoinhibitory motif. These plants were slightly smaller than wild-type plants (Figure 2) and had slightly elevated Tre6P levels (Figure 7A), a higher Tre6P:Suc ratio (Figure 7A), and higher levels of TCA cycle intermediates (Supplemental Figure 20), resembling the metabolic effects associated with overexpression of TPS (Yadav et al., 2014; Figueroa et al., 2016).
The putative Ser-252 phosphorylation site, identified by in vitro phosphorylation experiments on TPS1-derived peptides (Glinski and Weckwerth, 2005), is in the glucosyltransferase domain. We observed no obvious developmental or metabolic phenotypes in the TPS1[S252A] line, where the putative phosphorylation site was absent, or in the phospho-mimic TPS1[S252D] line (Supplemental Figures 4, 11A, and 14). Searches in the PhosPhAt 4.0 database (Heazlewood et al., 2008; http://phosphat.uni-hohenheim.de/) indicated that phosphorylation of Ser-252 has never been experimentally observed in Arabidopsis plants, and it is also worth noting that this residue is absent from the other class I TPS isoforms in Arabidopsis TPS2 and TPS4 and is not universally conserved in TPS1 orthologs in other plant species, especially not in the grasses (Poaceae). Thus, we conclude that Ser-252 does not play a major role in the regulation of TPS1 activity in vivo.
The TPP-like C-terminal domain of TPS1 appears to be very important, as the removal of this domain led to severe growth, developmental, and metabolic phenotypes in the TPS1[∆C] lines (Figures 2, 3, and to 6 to 8; Supplemental Figures 4, 6, 11 to 14, and 16F). These phenotypes were essentially replicated in the TPS1[A119W] lines, which have a single-amino acid substitution in the catalytic domain. Both lines had markedly elevated levels of Tre6P (Figures 6 and 7), a trait that is usually associated with early flowering (Schluepmann et al., 2003) and low Suc content (Yadav et al., 2014). However, neither of these lines flowered and they both accumulated unprecedentedly high levels of Suc, indicating that increased Tre6P is not the only, or even the prime, reason for their phenotypic defects. Curiously, in both cases, the phenotypes were less severe when the N-terminal domain was also removed. These observations demonstrate that all three domains of TPS1 are required for its full functionality and indicate that there are complex interactions between the three domains (Figure 9C).
Sequence analysis and searches of the PhosPhAt 4.0 database identified two potential phosphorylation sites (Ser-826 and Ser-941; Wang et al., 2013; Roitinger et al., 2015) and a putative sumoylation site (Lys-902) in the C-terminal domain of TPS1. The putative sumoylation site and surrounding motif, S895⋅W⋅N⋅V⋅L⋅D⋅L⋅(sumoK)⋅G⋅E⋅N⋅Y⋅F⋅S⋅C909, are highly conserved among class I TPS proteins in plants (Supplemental Figure 19), including the streptophyte alga Klebsormidium flaccidum from the base of the land plant lineage, indicating an ancient origin and retention of this putative regulatory site during plant evolution. These observations indicate that there is considerable scope for posttranslational regulation of TPS1 within the C-terminal domain of the protein (Figure 9B), which will be lost when the C-terminal domain is removed. The importance of this putative sumoylation site (Lys-902) and/or distal phosphorylation site (Ser-941) was corroborated by the severe morphological and metabolic phenotypes of the TPS1[ΔC895-942] truncation line (Figures 3 and 6), in which both sites were removed. Further site-directed mutagenesis experiments will be required to confirm these putative regulatory sites and investigate their individual physiological significance.
It has long been known that some enzymes are prone to catalytic infidelity, due to substrate promiscuity or mistakes during catalysis. Rubisco (EC 4.1.1.39) is a classic example in plants, not only showing high activity with the wrong substrate (oxygen) but also generating a range of by-products that can act as dead-end inhibitors of the enzyme and need to be removed by the specialized repair enzyme Rubisco activase (Parry et al., 2008). In recent years, it has been recognized that many other plant enzymes make mistakes during catalysis (Hanson et al., 2016). It is intriguing that the three tps1-1 complementation lines with the most severe phenotypes—TPS1[∆C], TPS1[ΔC895-942], and TPS1[A119W]—all contained relatively high levels of two unidentified disaccharide-monophosphates (Supplemental Figure 18) that are present in only trace amounts in wild-type Arabidopsis plants (Figueroa and Lunn, 2016). TPS1[∆N∆C] (±GFP) and TPS1[∆N A119W] plants also contained elevated levels of the two unknown disaccharide-monophosphates, but to a lesser extent than in TPS1[∆C], TPS1[ΔC895-942], or TPS1[A119W] (Supplemental Figure 18), and their developmental and metabolic phenotypes were correspondingly milder.
A speculative explanation for the increased levels of the two unknown compounds in these lines is that the catalytic fidelity of the TPS1 enzyme is compromised by the absence of the C-terminal domain or by the presence of the A119W point mutation in the catalytic domain. Errors during catalysis may give rise to unnatural Tre6P isomers (e.g., α,β-1,1-Tre6P or β,β-1,1-Tre6P) instead of the authentic product, α,α-1,1-Tre6P. Alternatively, binding of inappropriate substrates, such as UDP-galactose, in the active site could generate heterosidic analogs of Tre6P (e.g., α,α-1,1-galactosyl-glucose 6-phosphate). Such stereoisomers and analogs of Tre6P could be potent ligands for Tre6P binding proteins, thereby disrupting Tre6P signaling pathways. Our data provide circumstantial evidence that Tre6P signaling is indeed disrupted in the lines with elevated levels of the two unidentified disaccharide-monophosphates. The TPS1[∆N∆C] and TPS1[∆N A119W] lines were both late-flowering (Figure 2) and the TPS1[∆C], TPS1[∆C895-942], and TPS1[A119W] plants never flowered, despite having elevated levels of Tre6P, which would otherwise be expected to trigger early flowering (Schluepmann et al., 2003). In general, the severity of their phenotypes was correlated with the levels of the two unknown compounds. Production of such aberrant Tre6P analogs might explain why Arabidopsis TPS1[A119W] could not complement the yeast tps1Δ mutant, even though this form of the enzyme retains the capacity for Tre6P synthesis (see above). Further studies are needed to establish the identities of the two unknown compounds and determine if they have any physiological role in modulating Tre6P signaling in wild-type plants.
Role of TPS1 in the Sucrose-Tre6P Nexus
The sucrose-Tre6P nexus model was originally founded on two observations: (1) Tre6P levels are highly correlated with Suc during diurnal fluctuations in rosettes and in response to exogenous Suc supplied to seedlings (Lunn et al., 2006); and (2) imposed changes in the level of Tre6P, by overexpression of TPS or TPP, lead to opposite effects on Suc levels (Yadav et al., 2014). It is poorly understood how changes in Suc content lead to parallel changes in the level of Tre6P. Inhibitor studies suggested that the Tre6P response to Suc is not dependent on de novo transcription but depends on translation (Yadav et al., 2014). However, polysome-loading and immunoblotting experiments showed that Suc does not upregulate the production of TPS1 itself, so it was proposed that Suc might induce the synthesis of a putative regulatory protein that activates TPS1 (Yadav et al., 2014). Phosphorylation and other posttranslational modifications are obvious potential mechanisms for the regulation of TPS1 by other proteins. Our results indicate that phosphorylation of Ser-252 is not a major factor in the regulation of TPS1, but the putative phosphorylation (Ser-826 and Ser-941) and sumoylation (Lys-902) sites in the C-terminal domain of TPS1 (Figure 9B) are worth investigating for their regulatory potential in future experiments.
Our results show that the catalytic activity of TPS1 is strongly influenced by its C-terminal TPP-like domain, which retains some of the active site residues involved in substrate binding in catalytically active TPPs but not the critical Asp residue that forms the phosphoacyl intermediate during catalysis (Lunn, 2007). Suc6P phosphatase (SPP) catalyzes the analogous reaction to TPP in the pathway of Suc biosynthesis. SPP is competitively inhibited by Suc and even more strongly by trehalose, both of which bind to a glucose binding site in the “cap” domain of the enzyme (Fieulaine et al., 2005, 2007). By analogy, we speculate that Suc might bind to the TPP-like domain of TPS1 via a similar mechanism and activate TPS activity allosterically or by promoting interaction with a putative protein activator (Protein X; Figure 9C; Yadav et al., 2014). There is also potential for Tre6P to inhibit TPS1 in a competitive manner by binding in the active site or allosterically by binding to the TPP-like domain (Figure 9C). In the latter scenario, binding of a putative inhibitory protein (Protein Y; Figure 9C) in the presence of Tre6P is a further possibility. Activity and ligand binding assays of recombinant TPS1, including truncated or mutated versions, will be required to explore these hypothetical mechanisms for the regulation of TPS1.
In most of the tps1-1 complementation lines transformed with variants of TPS1, Tre6P tracked the endogenous fluctuations in leaf Suc levels during the diurnal cycle, and these two metabolites were highly correlated (Figure 7). The two exceptions were the TPS1[∆C] and TPS1[A119W] lines, where Tre6P and Suc were poorly correlated. The tight linkage between Suc and Tre6P also appeared to be broken in leaves of OtsA-transformed plants, with Tre6P barely changing during the light-dark cycle, while the diurnal fluctuation in Suc was slightly greater than in wild-type plants (Figure 7A). The greater accumulation of Suc in rosettes of OtsA lines during the day could be indicative of less Suc export to the roots, potentially explaining the poor root growth of these plants (Supplemental Figure 5). Another possibility is that the expression of otsA in the roots, instead of TPS1, disrupts the operation of the sucrose-Tre6P nexus in the roots, potentially delivering incorrect signals on the availability of Suc, which compromises root growth. We also cannot exclude the possibility that TPS1 has some other function in roots that is not directly linked to its catalytic activity. Regulation of maize inflorescence branching by the maize RAMOSA3 and ZmTPP4 proteins is uncoupled from their catalytic activities (Claeys et al., 2019), setting a precedent for Tre6P-metabolizing enzymes to have moonlighting functions. Overall, these observations point to OtsA acting as an adequate source of Tre6P when expressed in planta but lacking some functionalities of TPS1.
In wild-type Col-0 and TPS1 control seedlings, Tre6P was strongly responsive to exogenous supply of Suc, rising rapidly, after a short lag, to reach levels that were up to 140-fold higher than in carbon-starved seedlings (Figure 8; Lunn et al., 2006; Yadav et al., 2014). Tre6P also rose after Suc addition to TPS1[∆N∆C], TPS1[∆C], and OtsA seedlings but less strongly than in the Col-0 and TPS1 controls (Figure 8). It is likely that simple mass-action effects (i.e., increased availability of the UDP-glucose and Glc6P substrates for Tre6P synthesis) made some contribution to these responses, although previous studies showed only weak correlations between Tre6P and UDP-glucose or Glc6P in wild-type Col-0 seedlings (Yadav et al., 2014). Collectively, these results indicate that the sucrose-Tre6P nexus relationship is more or less fully restored in the TPS1, SV40-TPS1, and TPS1[L27P] lines, partially restored in the TPS1[∆N] and TPS1[∆N∆C] lines, but dysfunctional in the TPS1[∆C], TPS1[A119W], and OtsA lines.
This pattern does not correlate with the severity of the phenotypes of the complementation lines, questioning the physiological significance of the sucrose-Tre6P nexus relationship. This is best exemplified by the contrast between the OtsA plants, with near-wild-type phenotypes except for their shorter roots, and the TPS1[∆C] and TPS1[A119W] plants, which were severely dwarfed, nonflowering, and had very different metabolite profiles compared with wild-type plants. However, caution is needed in interpreting the strength of the correlation between Tre6P and Suc when based on whole-rosette or whole-seedling measurements of these metabolites. The localization of TPS1 in specific tissues and cell types (e.g., phloem and guard cells) suggests that the distribution of Tre6P at the whole-plant level may be more heterogeneous than previously assumed (Martins et al., 2013) and responsive to changes in Suc levels only in specific cells, such as phloem parenchyma, companion cells, sieve elements, and guard cells. Subcellular compartmentation adds a further potential layer of complexity; the nuclear localization of TPS1 suggests that Tre6P is responsive to Suc levels in the nucleocytosolic compartment of those cells where TPS1 is expressed. However, this probably constitutes a minor fraction of the total Suc in the plant, as there are large pools of Suc in vacuoles, and some of the Suc in rosettes is present in the apoplast or in the mesophyll cell chloroplasts (Lunn, 2016). Indeed, given the differences in the cellular and subcellular distributions of Tre6P and Suc, it is quite remarkable that such strong correlations between Tre6P and Suc are ever observed at the whole-plant level. Following this line of thought, it is possible that the sucrose-Tre6P nexus relationship is still operating in critical cell types in otsA lines, accounting for the relatively mild phenotypes of these plants, even though at the whole-plant level Tre6P and Suc are not well correlated. Another possibility is that the severe phenotypes of the TPS1[∆C] and TPS1[A119W] lines (and presumably also the TPS1[ΔC895-942] line) are primarily due to disruption of Tre6P signaling processes by the unknown disaccharide-monophosphates that are particularly abundant in these plants, rather than a complete breakdown in the sucrose-Tre6P nexus. In this scenario, any rise in Suc levels may still drive an increase in Tre6P. However, if the negative feedback effect of Tre6P was blocked by the unknown disaccharide-monophosphates, Suc would continue to accumulate, driving Tre6P higher as well. In other words, the plants would appear to be “blind” to Tre6P, failing to trigger the usual processes that lower Suc levels when Tre6P is high as well as the developmental responses (e.g., early flowering) that are typically observed in plants with high Tre6P. At present, it is unclear whether the high Suc and Tre6P levels in the dwarfed TPS1[∆C], TPS1[ΔC895-942], and TPS1[A119W] lines are due to a disruption of Suc export from the leaves to the roots, a disruption of the negative feedback loop in the sucrose-Tre6P nexus, or a combination of both.
In conclusion, the sucrose-Tre6P nexus model has provided a conceptual framework for investigating the function of Tre6P in plants and plausible explanations for the impact of manipulating Tre6P on plant metabolism and development. While it might be premature to abandon this model completely, the results of this study show that refinement is needed. The model postulates that Tre6P functions as both a signal and a negative feedback regulator of Suc levels. Given the diverse types of cells where TPS1 and, by inference, Tre6P are located, we need to consider the possibility that the relative importance of these two functions, although connected, may differ between tissues and cell types. In some parts of the plant and some developmental stages, homeostatic regulation of Suc levels might be the dominant role of Tre6P, while in others, signaling of Suc availability for growth might be more important. Therefore, in future experiments, it will be crucial to target manipulations of Tre6P to individual cell types and developmental stages to understand its specific functions, as exemplified by the work of Nuccio et al. (2015) in improving drought tolerance in maize.
METHODS
Materials
The Arabidopsis (Arabidopsis thaliana) Col-0 tps1-1 mutant (Eastmond et al., 2002) was kindly provided by Ian Graham (University of York). Proofreading Q5 High-Fidelity DNA polymerase, OneTaq DNA polymerase, T4 DNA ligase, and Q5 Site-Directed Mutagenesis Kit were obtained from New England Biolabs. All other enzymes for molecular cloning were obtained from Thermo Fisher Scientific. NucleoSpin Plasmid EasyPure, NucleoSpin Gel, and PCR Clean-up kits were obtained from Macherey-Nagel.
Molecular Cloning
Arabidopsis genomic DNA was isolated using the cetyl trimethylammonium bromide method (Doyle and Doyle, 1987) with elution of DNA in 100 μL of double-distilled water. All constructs were based on the pGreenII binary plasmid (Hellens et al., 2000), with the phosphino thricin N-acetyltransferase gene replaced by the nptII gene to allow selection of plant transformants with kanamycin. The Arabidopsis TPS1 (At1g78580) gene was amplified in sections from Arabidopsis Col-0 genomic DNA by PCR using Q5 High-Fidelity DNA polymerase and reassembled in the pGreenII[nptII] plasmid as follows: (1) the 3′-UTR and terminator region were ligated into the XmaI/HindIII sites; (2) the promoter region, including the 5′-UTR, was added in two parts using the KpnI/ScaI and ScaI/SpeI restriction sites (with ScaI naturally occurring in the TPS1 promoter; Figure 1C); and (3) the genomic protein-coding region, including all introns, was inserted into the SpeI/XmaI sites. A StuI restriction site was added after the start codon and an ApaI site was added before the stop codon, allowing removal of the N- and C-terminal domains by naturally occurring StuI and ApaI sites in the TPS1 sequence (Figures 1C and 1D). GFP, GUS, and SV40 tags were inserted at the N terminus using a PfoI site created by the addition of the StuI site to the TPS1 sequence (Figure 1C). For C-terminal GFP/GUS fusions of the wild-type and SV40-tagged versions of the full-length TPS1, the GFP/GUS coding sequences were fused to a DNA fragment corresponding to the TPS1 C-terminal domain in vitro (matching overlapping ends were generated by BstXI) and insertion of the resulting C terminus-GFP/GUS fusion DNA product back into the ApaI sites (Figure 1D). For C-terminal deletion constructs, the TPS1 fragment encoding the C terminus was replaced with the GFP sequence using the ApaI sites (Figure 1D). Site-directed mutagenesis was performed using the fully assembled pGreenII[nptII]/TPS1 plasmid as a template, with specific mutations being generated by PCR using a Q5 Site-Directed Mutagenesis Kit (New England Biolabs; Figure 1E). The otsA gene from Escherichia coli (strain K12) was amplified from E. coli DNA and inserted into the TPS1pro:TPS1term expression cassette using the SpeI/XmaI sites (Figure 1F). Oligonucleotide primers used for cloning are shown in Supplemental Table 1.
Arabidopsis Transformation and Cultivation
Constructs were introduced into heterozygous TPS1/tps1-1 plants (Col-0 background) by Agrobacterium tumefaciens (strain GV3101 pMP90; Koncz and Schell, 1986)-mediated transformation using the floral dip standard protocol (Clough and Bent, 1998). Primary transformants were selected by sowing surface-sterilized seeds on half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) containing 15 mg/L glufosinate, 50 mg/L kanamycin, and 0.05% (v/v) Plant Preservative Mixture (Plant Cell Technology), stratified for 3 d at 4°C, and then transferred to 22°C. After 7 d, surviving seedlings were transferred to soil (in 6-cm pots, one seedling per pot). The plants were selfed and T2 progeny were screened by selection on glufosinate and kanamycin. The genotype at the TPS1 locus in individual plants was determined by PCR on genomic DNA using the oligonucleotide primers shown in Supplemental Table 1. We used segregation analysis of the T3 progeny on glufosinate and kanamycin to identify lines that were homozygous for both the tps1-1 locus and the introduced pGreenII[nptII]-derived transgene. Supplemental Table 2 lists all the genotypes generated in the course of this study.
For phenotyping and metabolite analyses, plants were grown in a 1:1 mixture of soil (Stender) and vermiculite, in controlled-environment chambers with either 16-h (long days) or 12-h (equinoctial) photoperiods, day/night temperatures of 22/18°C, and 150 to 160 µmol m−2 s−1 irradiance provided by fluorescent lights (see Annunziata et al. [2017] for light spectrum). Flowering time was determined as bolting time (inflorescence stem size ≥ 0.5 cm) counted as days after sowing or as total leaf number (rosette + cauline leaves). For metabolite analysis, rosettes were harvested in the ambient growth conditions by rapid quenching in liquid nitrogen, and two or three plants were pooled per sample. See the description of individual experiments for the age of the plants and time of harvesting.
For Suc-addition experiments on seedlings, plants were grown axenically on half-strength Murashige and Skoog medium agar plates in constant light (150 μmol m−2 s−1 irradiance) at 22°C. Ten-day-old seedlings were placed in the dark for 2 d to exhaust their carbon reserves and then transferred to 150-mL Erlenmeyer flasks containing 30 mL of culture medium containing 15 mM Suc (Osuna et al., 2007). Whole seedlings were harvested at 30, 90, and 180 min after transfer to liquid medium. Nondarkened seedlings were used as nonstarved controls.
Immunoblotting
Proteins were extracted from aliquots (20 mg) of finely powdered tissue by mixing with 10 volumes of extraction buffer: 10% (v/v) glycerol, 0.5% (w/v) SDS, 0.01% (w/v) bromophenol blue, 0.06 M Tris-HCl, pH 6.8, 0.05% (v/v) β-mercaptoethanol, and heating at 95°C for 5 min. After centrifugation at 13,000g for 5 min, aliquots of the supernatant were separated on 10% (w/v) polyacrylamide gels containing 0.1% (w/v) SDS. Proteins were electroblotted onto 0.45-µm nitrocellulose membranes (Amersham Protran, GE Healthcare Life Sciences) and stained with 0.1% (w/v) Ponceau Red in 1% (v/v) acetic acid. The Ponceau Red-stained membrane was digitally scanned as a record of protein loadings. After destaining, membranes were blocked by incubation in 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.2% (v/v) Tween 20, and 0.2% (w/v) fat-free milk powder and incubated with purified rabbit α-TPS1 (N terminus; 1:1000 dilution; Yadav et al., 2014) antibodies in blocking solution at room temperature for 1 h. After washing, membranes were incubated with alkaline phosphatase-conjugated goat anti-rabbit IgG secondary antibodies (Promega) at a 1:7500 dilution in blocking buffer at room temperature for 1 h. After washing, immunoreactive proteins were detected colorimetrically by incubation with 0.15 mM 5-bromo-4-chloro-3-indolyl-phosphate-p-toluidine and 0.15 mM nitrotetrazolium-blue-chloride in 100 mM Tris-HCl, pH 8.5.
Metabolite Analysis
Frozen plant tissue was ground to a fine powder at liquid nitrogen temperature using a ball mill (Retsch). Aliquots (10 to 20 mg) of frozen tissue powder were extracted with chloroform-methanol as described by Lunn et al. (2006). After phase partitioning, the upper aqueous methanol phase was collected, evaporated to dryness, and dissolved in 350 μL of double-distilled water. The extract was filtered through MultiScreenHTS PCR Filter Plate membranes (Merck Millipore) to remove high-molecular-weight contaminants before analysis by LC-MS/MS or HPLC. The chloroform phase and insoluble residue from the interface were evaporated to dryness under vacuum, resuspended in 500 μL of 0.1 M NaOH, and solubilized by heating at 98°C for 30 min for determination of starch.
Soluble sugars were measured by LC-MS/MS as described by Fichtner et al. (2017). Tre6P, phosphorylated intermediates, and organic acids were measured by LC-MS/MS (Lunn et al., 2006), with modifications as described by Figueroa et al. (2016). Individual amino acids were measured by HPLC with precolumn derivatization with O-phthaldialdehyde and fluorescence detection (Watanabe et al., 2013). Starch was determined enzymatically (Hendriks et al., 2003).
Microscopy
GUS-Reporter Lines
To detect GUS activity, harvested plant material was placed in 50 mM sodium-phosphate buffer (pH 7), 5 mM K3[Fe(CN)6], 5 mM K4[Fe(CN)6], and 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (cyclohexyl-ammonium salt). After overnight incubation at 37°C in the dark, the tissue was destained by washing several times with 70% (v/v) ethanol. For sectioning, leaf tissue was dehydrated using an ethanol series and then embedded by incubation for 2 h at room temperature in a 1:1 mixture of ethanol and Technovit 7100 glycol methacrylate monomer (Kulzer). The embedded tissue was then transferred to infiltration solution (100 mL of Technovit 7100, 1 g of Hardener I) and incubated for 2 h at room temperature, then transferred and positioned in embedding solution (15 mL of infiltration solution, 1 mL of Hardener II). The embedding reagent was polymerized by overnight incubation of the samples at room temperature. Sections (4 µm thickness) were cut using a Leica Rotary Microtome RM2265 (Leica Biosystems), arrayed in distilled water on a glass microscope slide, and dried for ∼2 h at 42°C. Dried sections were observed with an Olympus BX-51 Epi-Fluorescence Microscope fitted with a DC View III camera and operated using CellSense software (Olympus). Nonsectioned plant material was examined using either the microscope described above or a Leica Stereomicroscope MZ12.5 fitted with a DC 420 camera and operated with LAS software (Leica Biosystems).
GFP-Reporter Lines
GFP expression was detected using a Leica TCS SP8 spectral laser scanning confocal motorized microscope operated with LAS X software (Leica Biosystems). Overlays were done using the image-processing package Fiji for ImageJ. Nuclei were stained by incubation of 8-d-old seedlings in 1× PBS containing 0.3% (v/v) Tween 20 and 2 mg/L DAPI for 10 min. Seedlings were washed twice in water before imaging.
Data Analysis and Statistics
Data plotting and statistical analysis were performed using R Studio version 1.0.163 with R version 3.6.1 (https://cran.r-project.org/) and the package ggplot2. Data were analyzed by an ANOVA-based posthoc comparison of means test using the multiple-comparison Fisher’s LSD test. The results of the statistical analyses are presented in the Supplemental Data Set. Heatmap analyses were performed with the heatmap.2 function (R package heatmaply) using the agglomeration method mcquitty for the hierarchical cluster analysis and the distance measure canberra for the computation of the distance matrix. Figures containing micrographs and other images were compiled using Microsoft PowerPoint 2010 or Adobe Illustrator. The presented images are representative of replicated samples within each experiment, with contrast, brightness, and other picture quality settings being adjusted in the same way for all comparable images.
Accession Numbers
Sequence data from this article can be found under the following accession numbers: TPS1 (At1g78580), TPS2 (At1g16980), TPS4 (At4g27550), and otsA (NC_000913). Accession numbers of TPS polypeptide sequences from Supplemental Figure 19 are shown in Supplemental Table 3.
Supplemental Data
Supplemental Figure 1. Expression atlas of TPS1 transcript in Arabidopsis.
Supplemental Figure 2. Complementation of the Arabidopsis tps1-1 mutant by β-GLUCURONIDASE (GUS) tagged TPS1 fusion proteins.
Supplemental Figure 3. Complementation of the Arabidopsis tps1-1 mutant by tagged, truncated or mutated forms of TPS1.
Supplemental Figure 4. Rosette morphology of tps1-1 complementation lines grown in long-day conditions.
Supplemental Figure 5. Root morphology of tps1-1 complementation lines grown in long-day conditions.
Supplemental Figure 6. Shoot morphology of tps1-1 complementation lines grown in long-day conditions.
Supplemental Figure 7. Expression pattern of the Arabidopsis TPS1 protein in reproductive tissues.
Supplemental Figure 8. Localization of the Arabidopsis TPS1 protein in the shoot apical meristem (SAM) and subtending stem .
Supplemental Figure 9. Localization of the Arabidopsis TPS1 protein in the inflorescence meristem and floral primordia.
Supplemental Figure 10. Localization of the Arabidopsis TPS1 protein in guard cells.
Supplemental Figure 11. Analysis of Tre6P and other metabolites in tps1-1 complementation lines grown in long-day conditions.
Supplemental Figure 12. Analysis of amino acids in tps1-1 complementation lines grown in long-day conditions.
Supplemental Figure 13. Hierarchical clustering analysis of Tre6P, Suc and other metabolite data of tps1-1 complementation lines in long days.
Supplemental Figure 14. Analysis of Tre6P and other metabolites in tps1-1 complementation lines grown in equinoctial conditions.
Supplemental Figure 15. Starch content of tps1-1 complementation lines grown in equinoctial or long-day conditions.
Supplemental Figure 16. Diurnal profiles of Tre6P and other metabolites in tps1-1 complementation lines grown in long-day conditions.
Supplemental Figure 17. Response of seedlings from tps1-1 complementation lines to exogenous Suc supply.
Supplemental Figure 18. Detection of two unidentified disaccharide-monophosphates in tps1-1 complementation lines.
Supplemental Figure 19. Alignment of the putative sumoylation site motif in the C-terminal domain of Arabidopsis TPS1 with orthologous sequences from class I TPS proteins in other plant species.
Supplemental Table 1. Oligonucleotide primers used for cloning and testing the zygosity of the tps1-1 mutant locus in transgenic Arabidopsis lines.
Supplemental Table 2. Genotypes generated during the course of this study.
Supplemental Table 3. Accession numbers of TPS polypeptide sequences shown in Supplemental Figure 19.
Supplemental Data Set. Statistical analysis.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Sven Roigk, Katrin Seehaus, and other members of the Greenteam for help with plant cultivation and transformation as well as Ralph Bock, Carlos Figueroa, Arun Sampathkumar, and Patrick Van Dijck for helpful discussions and advice. This work was supported by the International Max Planck Research School, Primary Metabolism and Plant Growth (Ph.D. scholarship to F.F.), and by the Max Planck Society (to J.J.O., R.F., M.W., U.K., R.H., M.S., and J.E.L.).
AUTHOR CONTRIBUTIONS
F.F., M.S., and J.E.L. conceived the project and designed the experiments; F.F. did the molecular cloning and generated the transgenic plant lines; F.F. and U.K. identified homozygous lines by PCR genotyping and immunoblotting; F.F and J.J.O. did the sectioning and microscopy; F.F. and R.F. measured metabolites by LC-MS/MS and enzymatic assays; M.W. and R.H. determined amino acids; F.F. did the statistical analysis and prepared the figures; F.F and J.E.L wrote the article with significant input from M.S.; all authors approved the final version.
Footnotes
Articles can be viewed without a subscription.
References
- Annunziata M.G., Apelt F., Carillo P., Krause U., Feil R., Mengin V., Lauxmann M.A., Köhl K., Nikoloski Z., Stitt M., Lunn J.E., Raines C.(2017). Getting back to nature: A reality check for experiments in controlled environments. J. Exp. Bot. 68: 4463–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S., Smith-Unna R.D., Boursnell C.M., Kopriva S., Hibberd J.M.(2014). Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant J. 78: 659–673. [DOI] [PubMed] [Google Scholar]
- Avonce N., Mendoza-Vargas A., Morett E., Iturriaga G.(2006). Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 6: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista R.A., Figueiredo D.D., Santos-González J., Köhler C.(2019). Auxin regulates endosperm cellularization in Arabidopsis. Genes Dev. 33: 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing O.E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W.R., Stitt M.(2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez M.A., Santos E., Flores C.L., Martínez-Zapater J.M., Salinas J., Gancedo C.(1998). Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J. 13: 685–689. [DOI] [PubMed] [Google Scholar]
- Cabib E., Leloir L.F.(1958). The biosynthesis of trehalose phosphate. J. Biol. Chem. 231: 259–275. [PubMed] [Google Scholar]
- Chelsky D., Ralph R., Jonak G.(1989). Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol. Cell. Biol. 9: 2487–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.Q., Qu X.Q., Hou B.H., Sosso D., Osorio S., Fernie A.R., Frommer W.B.(2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211. [DOI] [PubMed] [Google Scholar]
- Chen Q., Payyavula R.S., Chen L., Zhang J., Zhang C., Turgeon R.(2018). FLOWERING LOCUS T mRNA is synthesized in specialized companion cells in Arabidopsis and Maryland Mammoth tobacco leaf veins. Proc. Natl. Acad. Sci. USA 115: 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D.(2017). Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89: 789–804. [DOI] [PubMed] [Google Scholar]
- Claeys H., Vi S.L., Xu X., Satoh-Nagasawa N., Eveland A.L., Goldshmidt A., Feil R., Beggs G.A., Sakai H., Brennan R.G., Lunn J.E., Jackson D.(2019). Control of meristem determinacy by trehalose 6-phosphate phosphatases is uncoupled from enzymatic activity. Nat. Plants 5: 352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cookson S.J., Yadav U.P., Klie S., Morcuende R., Usadel B., Lunn J.E., Stitt M.(2016). Temporal kinetics of the transcriptional response to carbon depletion and sucrose readdition in Arabidopsis seedlings. Plant Cell Environ. 39: 768–786. [DOI] [PubMed] [Google Scholar]
- Delorge I., Figueroa C.M., Feil R., Lunn J.E., Van Dijck P.(2015). Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem. J. 466: 283–290. [DOI] [PubMed] [Google Scholar]
- Dos Anjos L., Pandey P.K., Moraes T.A., Feil R., Lunn J.E., Stitt M.(2018). Feedback regulation by trehalose 6-phosphate slows down starch mobilization below the rate that would exhaust starch reserves at dawn in Arabidopsis leaves. Plant Direct 2: e00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J., Doyle J.(1987). Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem. Bull. 19: 11–15. [Google Scholar]
- Eastmond P.J., van Dijken A.J.H., Spielman M., Kerr A., Tissier A.F., Dickinson H.G., Jones J.D.G., Smeekens S.C., Graham I.A.(2002). Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J. 29: 225–235. [DOI] [PubMed] [Google Scholar]
- Fichtner F., Barbier F.F., Feil R., Watanabe M., Annunziata M.G., Chabikwa T.G., Höfgen R., Stitt M., Beveridge C.A., Lunn J.E.(2017). Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J. 92: 611–623. [DOI] [PubMed] [Google Scholar]
- Fieulaine S., Lunn J.E., Borel F., Ferrer J.L.(2005). The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell. Plant Cell 17: 2049–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieulaine S., Lunn J.E., Ferrer J.L.(2007). Crystal structure of a cyanobacterial sucrose-phosphatase in complex with glucose-containing disaccharides. Proteins 68: 796–801. [DOI] [PubMed] [Google Scholar]
- Figueroa C.M., et al. (2016). Trehalose 6-phosphate coordinates organic and amino acid metabolism with carbon availability. Plant J. 85: 410–423. [DOI] [PubMed] [Google Scholar]
- Figueroa C.M., Lunn J.E.(2016). A tale of two sugars: Trehalose 6-phosphate and sucrose. Plant Physiol. 172: 7–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geelen D., Royackers K., Vanstraelen M., De Bus M., Inzé D., Van Dijck P., Thevelein J.M., Leyman B.(2007). Trehalose-6-P synthase AtTPS1 high molecular weight complexes in yeast and Arabidopsis. Plant Sci. 173: 426–437. [Google Scholar]
- Gibson R.P., Tarling C.A., Roberts S., Withers S.G., Davies G.J.(2004). The donor subsite of trehalose-6-phosphate synthase: Binary complexes with UDP-glucose and UDP-2-deoxy-2-fluoro-glucose at 2 A resolution. J. Biol. Chem. 279: 1950–1955. [DOI] [PubMed] [Google Scholar]
- Gibson R.P., Turkenburg J.P., Charnock S.J., Lloyd R., Davies G.J.(2002). Insights into trehalose synthesis provided by the structure of the retaining glucosyltransferase otsA. Chem. Biol. 9: 1337–1346. [DOI] [PubMed] [Google Scholar]
- Glinski M., Weckwerth W.(2005). Differential multisite phosphorylation of the trehalose-6-phosphate synthase gene family in Arabidopsis thaliana: A mass spectrometry-based process for multiparallel peptide library phosphorylation analysis. Mol. Cell. Proteomics 4: 1614–1625. [DOI] [PubMed] [Google Scholar]
- Gómez L.D., Baud S., Gilday A., Li Y., Graham I.A.(2006). Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J. 46: 69–84. [DOI] [PubMed] [Google Scholar]
- Gómez L.D., Gilday A., Feil R., Lunn J.E., Graham I.A.(2010). AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 64: 1–13. [DOI] [PubMed] [Google Scholar]
- Hanson A.D., Henry C.S., Fiehn O., de Crécy-Lagard V.(2016). Metabolite damage and metabolite damage control in plants. Annu. Rev. Plant Biol. 67: 131–152. [DOI] [PubMed] [Google Scholar]
- Haritatos E., Ayre B.G., Turgeon R.(2000). Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol. 123: 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harthill J.E., Meek S.E., Morrice N., Peggie M.W., Borch J., Wong B.H., Mackintosh C.(2006). Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J. 47: 211–223. [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Durek P., Hummel J., Selbig J., Weckwerth W., Walther D., Schulze W.X.(2008). PhosPhAt: A database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 36: D1015–D1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M.(2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Hendriks J.H., Kolbe A., Gibon Y., Stitt M., Geigenberger P.(2003). ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol. 133: 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper C.M., et al. (2017). Multiple marker abundance profiling: Combining selected reaction monitoring and data-dependent acquisition for rapid estimation of organelle abundance in subcellular samples. Plant J. 92: 1202–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Ham B.K., El-Shabrawi H.M., Alexander D., Zhang D., Ryals J., Lucas W.J.(2016). Proteomics and metabolomics analyses reveal the cucurbit sieve tube system as a complex metabolic space. Plant J. 87: 442–454. [DOI] [PubMed] [Google Scholar]
- Koncz C., Schell J.(1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396. [Google Scholar]
- Kosugi S., Hasebe M., Matsumura N., Takashima H., Miyamoto-Sato E., Tomita M., Yanagawa H.(2009). Six classes of nuclear localization signals specific to different binding grooves of importin α. J. Biol. Chem. 284: 478–485. [DOI] [PubMed] [Google Scholar]
- Leyman B., Van Dijck P., Thevelein J.M.(2001). An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci. 6: 510–513. [DOI] [PubMed] [Google Scholar]
- Lunn J.E.(2007). Gene families and evolution of trehalose metabolism in plants. Funct. Plant Biol. 34: 550. [DOI] [PubMed] [Google Scholar]
- Lunn J.E.(2016). Sucrose metabolism. In eLS. (Chichester, UK: John Wiley & Sons; ), pp. 1–9. [Google Scholar]
- Lunn J.E., Ashton A.R., Hatch M.D., Heldt H.W.(2000). Purification, molecular cloning, and sequence analysis of sucrose-6F-phosphate phosphohydrolase from plants. Proc. Natl. Acad. Sci. USA 97: 12914–12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn J.E., Delorge I., Figueroa C.M., Van Dijck P., Stitt M.(2014). Trehalose metabolism in plants. Plant J. 79: 544–567. [DOI] [PubMed] [Google Scholar]
- Lunn J.E., Feil R., Hendriks J.H., Gibon Y., Morcuende R., Osuna D., Scheible W.R., Carillo P., Hajirezaei M.R., Stitt M.(2006). Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 397: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M.C., et al. (2013). Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6-phosphate. Plant Physiol. 163: 1142–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitzel T., Radchuk R., McAdam E.L., Thormählen I., Feil R., Munz E., Hilo A., Geigenberger P., Ross J.J., Lunn J.E., Borisjuk L.(2019). Trehalose 6-phosphate controls seed filling by inducing auxin biosynthesis. bioRxiv 752915. [DOI] [PubMed] [Google Scholar]
- Mitra S.K., Walters B.T., Clouse S.D., Goshe M.B.(2009). An efficient organic solvent based extraction method for the proteomic analysis of Arabidopsis plasma membranes. J. Proteome Res. 8: 2752–2767. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F.(1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nguyen Ba A.N., Pogoutse A., Provart N., Moses A.M.(2009). NLStradamus: A simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics 10: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio M.L., Wu J., Mowers R., Zhou H.-P., Meghji M., Primavesi L.F., Paul M.J., Chen X., Gao Y., Haque E., Basu S.S., Lagrimini L.M.(2015). Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 33: 862–869. [DOI] [PubMed] [Google Scholar]
- Nunes C., O’Hara L.E., Primavesi L.F., Delatte T.L., Schluepmann H., Somsen G.W., Silva A.B., Fevereiro P.S., Wingler A., Paul M.J.(2013). The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiol. 162: 1720–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna D., Usadel B., Morcuende R., Gibon Y., Bläsing O.E., Höhne M., Günter M., Kamlage B., Trethewey R., Scheible W.R., Stitt M.(2007). Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 49: 463–491. [DOI] [PubMed] [Google Scholar]
- Parry M.A., Keys A.J., Madgwick P.J., Carmo-Silva A.E., Andralojc P.J.(2008). Rubisco regulation: A role for inhibitors. J. Exp. Bot. 59: 1569–1580. [DOI] [PubMed] [Google Scholar]
- Piazza A., Zimaro T., Garavaglia B.S., Ficarra F.A., Thomas L., Marondedze C., Feil R., Lunn J.E., Gehring C., Ottado J., Gottig N.(2015). The dual nature of trehalose in citrus canker disease: A virulence factor for Xanthomonas citri subsp. citri and a trigger for plant defence responses. J. Exp. Bot. 66: 2795–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M., De Smet I., Vandesteene L., Naudts M., Leyman B., Van Dijck P., Rolland F., Beeckman T., Thevelein J.M.(2009). Extensive expression regulation and lack of heterologous enzymatic activity of the class II trehalose metabolism proteins from Arabidopsis thaliana. Plant Cell Environ. 32: 1015–1032. [DOI] [PubMed] [Google Scholar]
- Rao N.R., Kumaran D., Seetharaman J., Bonanno J.B., Burley S.K., Swaminathan S.(2006). Crystal structure of trehalose-6-phosphate phosphatase-related protein: Biochemical and biological implications. Protein Sci. 15: 1735–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitinger E., Hofer M., Köcher T., Pichler P., Novatchkova M., Yang J., Schlögelhofer P., Mechtler K.(2015). Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol. Cell. Proteomics 14: 556–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia D., Lunn J.E.(2017). Transitory starch metabolism in guard cells: Unique features for a unique function. Plant Physiol. 174: 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Nagasawa N., Nagasawa N., Malcomber S., Sakai H., Jackson D.(2006). A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230. [DOI] [PubMed] [Google Scholar]
- Schluepmann H., Pellny T., van Dijken A., Smeekens S., Paul M.(2003). Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 6849–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U.(2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506. [DOI] [PubMed] [Google Scholar]
- Vandesteene L., Ramon M., Le Roy K., Van Dijck P., Rolland F.(2010). A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol. Plant 3: 406–419. [DOI] [PubMed] [Google Scholar]
- Van Dijck P., Mascorro-Gallardo J.O., De Bus M., Royackers K., Iturriaga G., Thevelein J.M.(2002). Truncation of Arabidopsis thaliana and Selaginella lepidophylla trehalose-6-phosphate synthase unlocks high catalytic activity and supports high trehalose levels on expression in yeast. Biochem. J. 366: 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken A.J., Schluepmann H., Smeekens S.C.(2004). Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol. 135: 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G., Aeschbacher R.A., Müller J., Boller T., Wiemken A.(1998). Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: Identification by functional complementation of the yeast tps2 mutant. Plant J. 13: 673–683. [DOI] [PubMed] [Google Scholar]
- Wahl V., Ponnu J., Schlereth A., Arrivault S., Langenecker T., Franke A., Feil R., Lunn J.E., Stitt M., Schmid M.(2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707. [DOI] [PubMed] [Google Scholar]
- Wang X., Bian Y., Cheng K., Gu L.-F., Ye M., Zou H., Sun S.S.-M., He J.-X.(2013). A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J. Proteomics 78: 486–498. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Balazadeh S., Tohge T., Erban A., Giavalisco P., Kopka J., Mueller-Roeber B., Fernie A.R., Hoefgen R.(2013). Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol. 162: 1290–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Asención Diez M.D., Figueroa C.M., Machtey M., Iglesias A.A., Ballicora M.A., Liu D.(2015). The crystal structure of Nitrosomonas europaea sucrose synthase reveals critical conformational changes and insights into sucrose metabolism in prokaryotes. J. Bacteriol. 197: 2734–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav U.P., Ivakov A., Feil R., Duan G.Y., Walther D., Giavalisco P., Piques M., Carillo P., Hubberten H.M., Stitt M., Lunn J.E.(2014). The sucrose-trehalose 6-phosphate (Tre6P) nexus: Specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 65: 1051–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z., Keereetaweep J., Liu H., Feil R., Lunn J.E., Shanklin J.(2018). Trehalose 6-phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell 30: 2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Primavesi L.F., Jhurreea D., Andralojc P.J., Mitchell R.A., Powers S.J., Schluepmann H., Delatte T., Wingler A., Paul M.J.(2009). Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 149: 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., Ren J.(2014). GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42: W325–W330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Zhang W., Stanley B.A., Assmann S.M.(2008). Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20: 3210–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]