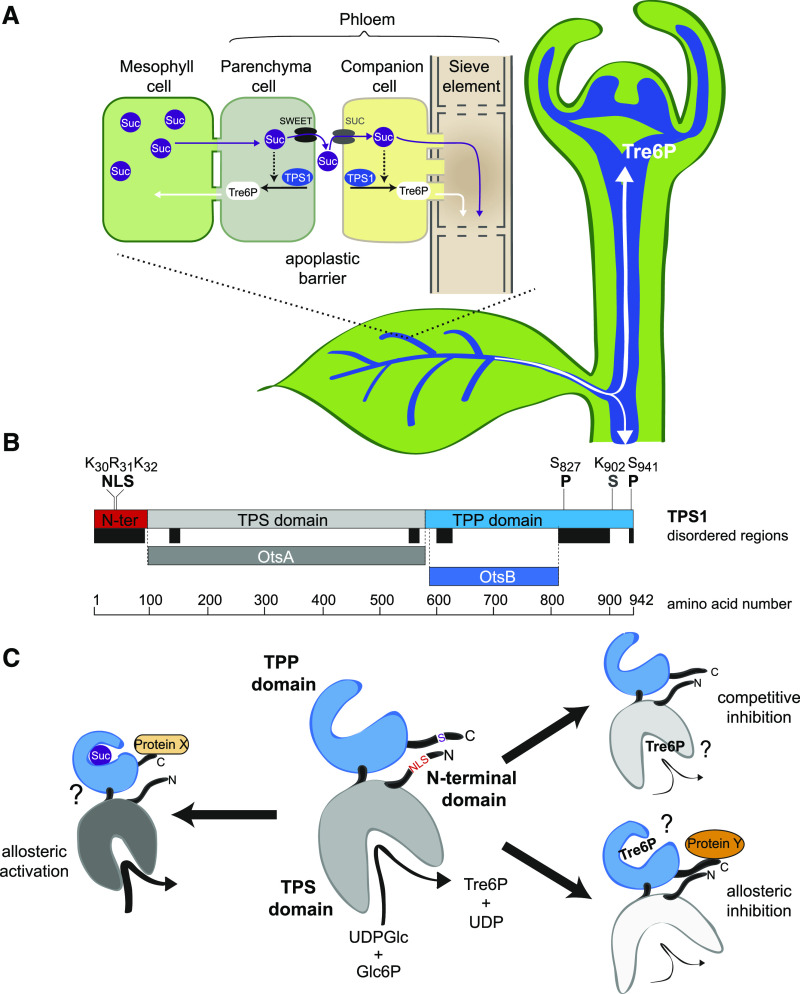
Figure 9.
Localization of TPS1 in the Phloem-Loading Zone and Potential Regulation of TPS1 Activity in Arabidopsis.
(A) Schematic diagram of TPS1 expression in the phloem parenchyma (bundle sheath) and companion cell/sieve element complex in leaf vascular bundles and in the SAM. Tre6P synthesized in the phloem parenchyma can diffuse symplastically into mesophyll cells to regulate photoassimilate partitioning (day) and transitory starch turnover (night) according to the demand for Suc. Tre6P synthesized in the companion cells can diffuse symplastically into the sieve elements and be transported around the plant, potentially acting as a systemic signal of Suc availability from source leaves.
(B) TPS1 has three domains: an N-terminal domain (red), a catalytic glucosyltransferase (TPS) domain (gray), and a TPP-like domain (blue). Important amino acid residues for the NLS, phosphorylation sites (P), and the sumoylation site (S) identified in this study are indicated. Colored bars below TPS1 indicate predicted disordered regions (black) and regions that are homologous with bacterial TPS (OtsA; dark gray) and TPP (OtsB; dark blue) enzymes.
(C) Schematic representation of TPS1 structure, based on the known structures of bacterial OtsA (Gibson et al., 2002, 2004) and OtsB (Rao et al., 2006) proteins, showing potential mechanisms of regulation by Suc and Tre6P. Left, potential allosteric activation of TPS1 by Suc binding to the C-terminal domain, possibly involving interaction with a putative activator protein (Protein X; Yadav et al., 2014). Right (top), competitive inhibition by Tre6P binding in the active site of TPS1. Right (bottom), potential allosteric inhibition of TPS1 by Tre6P binding to the C-terminal domain, conceivably involving interaction with a hypothetical inhibitory protein (Protein Y). These scenarios are not mutually exclusive and would allow a tight control of TPS1 activity based on the sucrose:Tre6P ratio in a given cell. The intracellular localization of TPS1 might also affect its activity and is potentially regulated by modification or interactions of other proteins with the NLS in the N-terminal domain.