Figure 1.
Genome-Wide Structural Variant Discovery in an Arabidopsis Population.
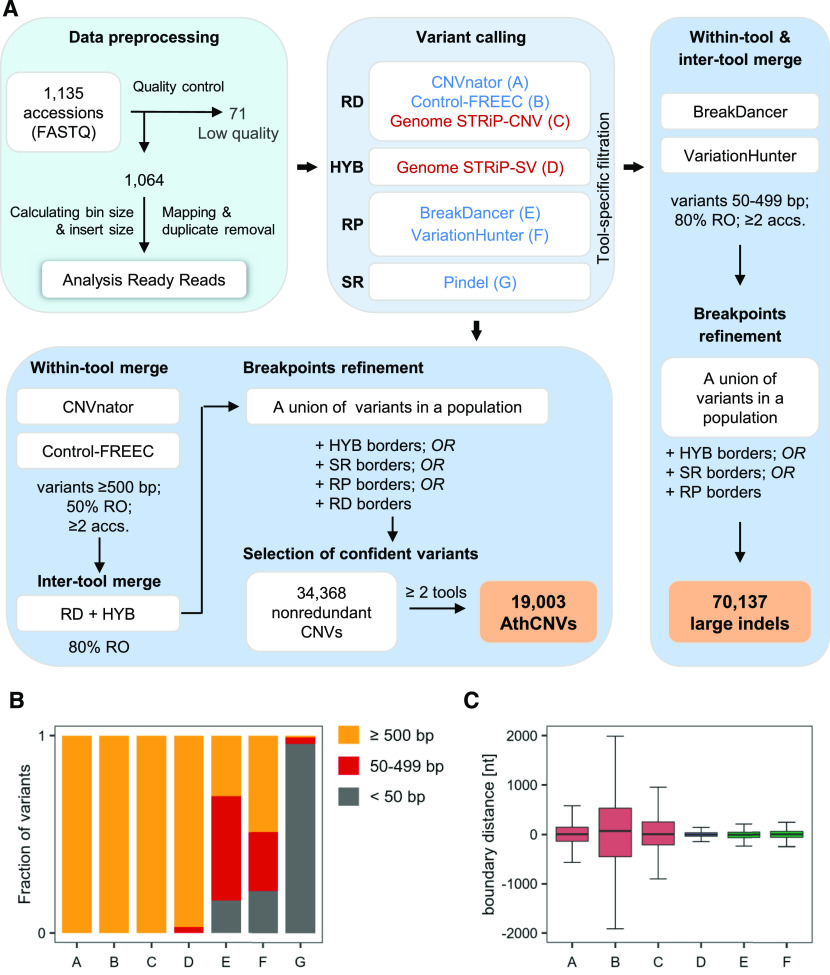
(A) Variant identification pipeline. The analysis involved three main stages: data preprocessing, variant calling, and merging and filtering. Variants were called with seven different tools, based on read depth (RD), read pair (RP), split read (SR), or hybrid (HYB) approach, in individual samples (blue labels) or in the entire population (red labels). The last stage depended on variant length. RO, reciprocally overlapping each other.
(B) Fraction of variants of different size ranges identified by individual callers.
(C) Comparison of the boundaries set by the callers for variants ≥500 bp reciprocally overlapping each other by 80%. Pindel-derived coordinates served as a reference since this tool reports variants at single-nucleotide resolution. Boxplots show median (inner line) and inner quartiles (box). Whiskers extend to the highest and lowest values no greater than 1.5 times the inner quartile range. nt, nucleotides.