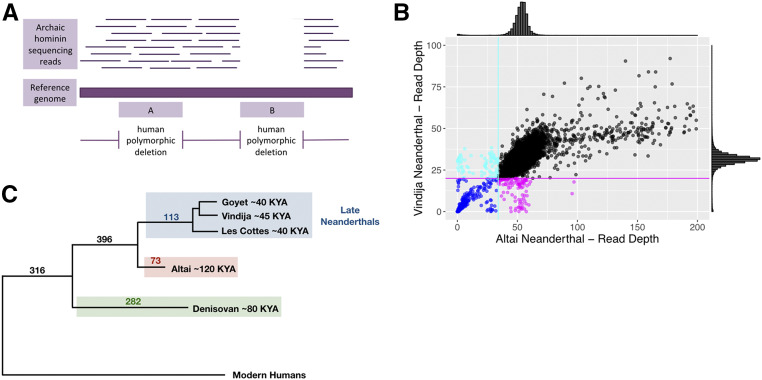
Figure 3.
Genotyping deletion variants found polymorphic in modern humans for ancient hominin genomes. (A) Schematic representation of genotyping the Neanderthal genomes for the human-polymorphic deletion variants included in the 1000 Genomes Project data set (Phase III). Horizontal thin lines show the raw sequencing reads for ancient hominin genomes. A horizontal thick line represents the human reference genome on which the raw sequencing reads for the ancient hominin genome are mapped. Regions found deleted in modern humans are shown in the lower part of the figure with pink shaded boxes. Box A represents an example region deleted in some modern human genomes where the ancient hominin genome carries raw sequence reads. Box B represents an example region deleted in some modern human genomes where the ancient hominin genome lacks raw sequence reads. (B) Raw read depth distribution of Neanderthal genomes for the regions found polymorphically deleted in humans. Histograms above the x- and y-axes show the read-depth distribution for the Altai and Vindija Neanderthal genomes. A normal distribution with the mean and SD of the empirical read depth distribution was fitted on the empirical read-depth distribution for the two Neanderthal genomes. Cyan and magenta lines show the 0.01 quantile values of normal distributions (34 and 20 for Altai and Vindija Neanderthal, respectively). Cyan and magenta points below these lines are considered as deleted in the Altai and Vindija Neanderthal genomes, respectively. Blue points show the regions where both Neanderthal genomes are genotyped as deleted. Black points show the regions where both Neanderthal genomes carry the intact sequences. (C) A schematic tree of ancient hominins and modern humans. The numbers of deletion variants shared with each branch of ancient hominins are shown on the corresponding branches.