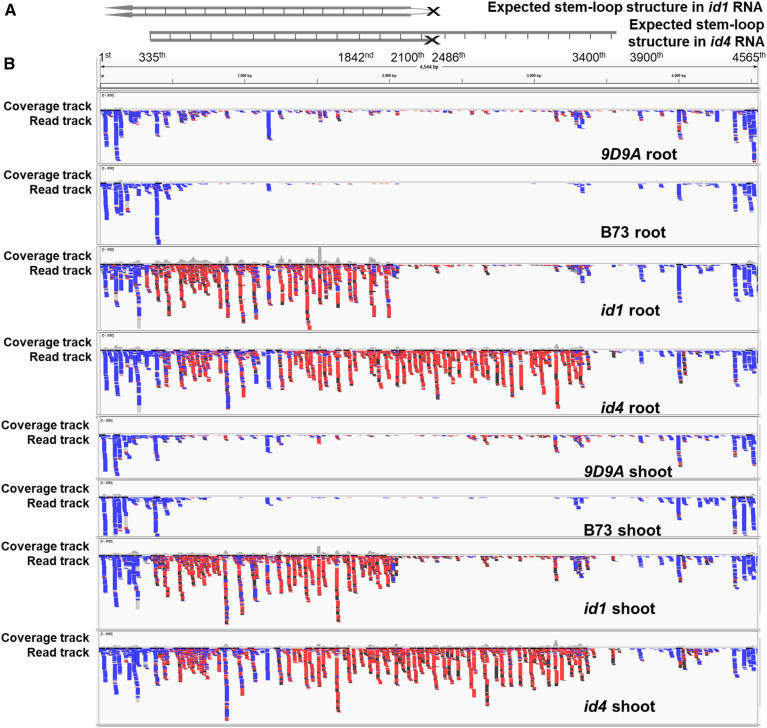
Figure 8.
Detection and mapping of small RNAs in p1-ww-id1 and p1-ww-id4. (A) The expected stem-loop structures in p1-ww-id1 and p1-ww-id4. The schematic structures include a ruler with corresponding nucleotide position in Ac. The loop of p1-ww-id4 is open for better presentation of the size. The transcription of Composite Insertion of p1-ww-id1 is driven by the host promoter, and the two inverted fAc fragments in p1-ww-id1 are relatively symmetrical (2100–2486 nt of each indicated by Southern blot), predicting a hairpin structure in the dsRNA contains first ∼ (2100th–2486th) nucleotides of Ac. The transcription of Composite Insertion of p1-ww-id4 is driven by the Ac promoter, and thus the transcript starts from 335th nucleotide of Ac. The inverted fAc fragments in p1-ww-id4 differ in size, with one fAc fragment of 1842–2486 nt and the other fAc of 3400–3900 nt indicated by Southern blot. This asymmetrical structure should give rise to a transcript with a stem containing 335th ∼ (1842nd–2486th) nucleotides of Ac and a loop containing (1843rd–2487th) ∼ (3400th–3900th) nucleotides of Ac in p1-ww-id4. (B) Small RNAs mapped to Ac are visualized by the Integrative Genomics Viewer (Robinson et al. 2011). The y axis of the coverage track was standardized to the same scale for each sample. Colors indicate size of siRNAs: black, 21 nt; red, 22 nt; blue, 24 nt.