Figure 1. p38γ is elevated in CTCL and is important for viability.
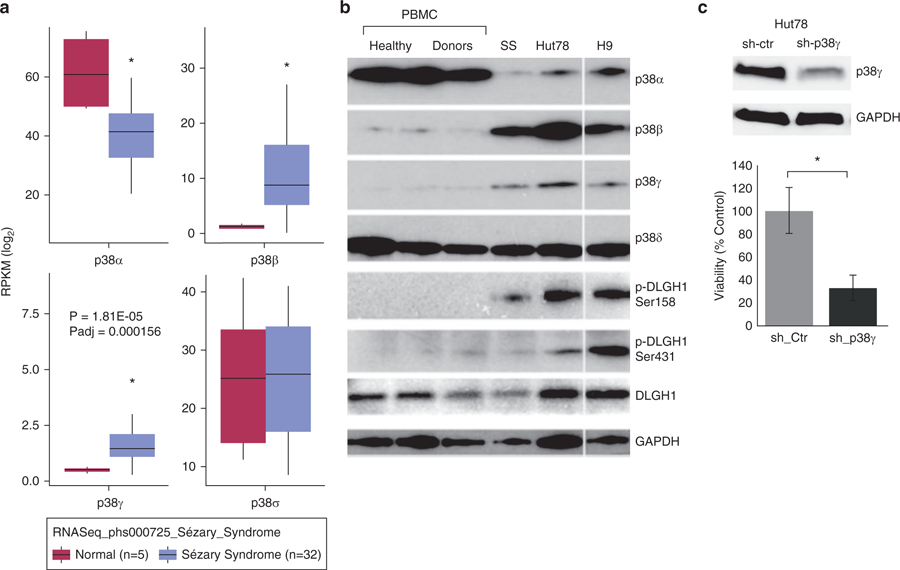
(a) RNA sequencing database phs000725 was downloaded from the dbGAP database with permission. Differential expression analysis of four p38 isoforms between CD4+ T cells of healthy donors (n = 5, pink) and that of patients with Sézary syndrome (n = 32, blue. *P < 0.005) using DESeq 2 package in R. Y-axis indicates log fold changes of expressed genes’ RPKM (reads per kilobase of transcript per million mapped reads). (b) Western blot was used to visualize protein expression of indicated p38 isoforms in peripheral blood mononuclear cells (PBMCs) of healthy donors (n = 3) and one patient with Sézary syndrome (SS), as well as whole cell lysates of Hut78 and H9 CTCL cell lines. White space indicates that the blotting of H9 sample are from different parts of the same gel with the same exposure. DLGH1 is a downstream target of p38γ kinase activity, indicated by phosphorylation at the 158 residue (p-DLGH1 Ser158); phosphorylation at the unrelated 431 residue (p-DLGH1 Ser431) is shown as a control. GAPDH is a control for protein loading. (c) Hut78 cells were transduced with lentiviral particles that harbor shRNA against p38γ or control shRNA. (Top) Western blot was used to visualize protein expression of p38γ. GAPDH is a control for protein loading. (Bottom) Cell viability was measured by trypan blue exclusion, and data are presented as a percentage of control-treated cells. Three replicates were performed for each sample. *P < 0.05. CTCL, cutaneous T-cell lymphoma; Ctr, control; sh, short hairpin RNA.
