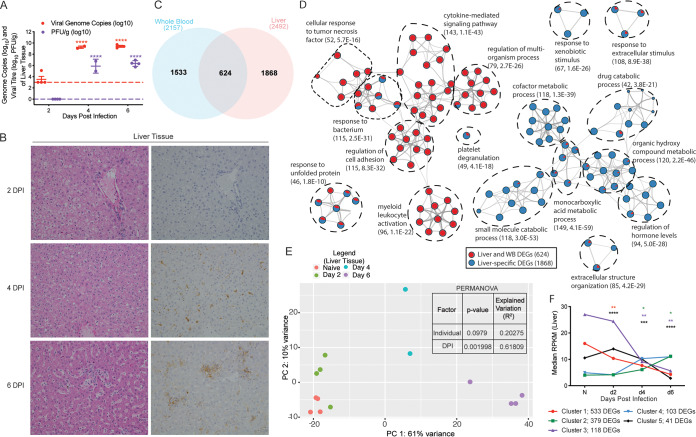
FIG 1.
Makona infection results in significant histological and transcriptional changes in the liver. (A) Mean ± standard deviation (SD) of EBOV infectious virus titer and genome copy numbers in liver were quantified using plaque assay and RT-qPCR for each animal at each time point. Significance was determined using one-way ANOVA with Dunnett’s multiple-comparison test relative to the limit of detection, which is represented as a dashed line. ****, P < 0.0001. (B) Hematoxylin and eosin staining (left) and immunohistochemistry staining (right) using EBOV-specific anti-VP40 antibody of liver at 2, 4, and 6 DPI (brown indicates reactivity); magnification ×20. (C) Venn diagram comparing differentially expressed genes (DEGs) detected in whole blood (WB) and liver pooled from all the time points analyzed. (D) Network image showing functional enrichment of DEGs detected in both liver and WB, and liver only generated using Metascape and visualized using Cytoscape. Each circle represents a theme of highly related gene ontology (GO) terms. The edges (lines) link similar GO terms, and the thickness of the line indicates the relatedness of the GO terms. Each GO term is shown as a pie chart that depicts the relative contribution of DEGs detected in both liver and WB (red) and liver only (blue). (E) Principal component analysis (PCA) of liver-specific DEGs over time. (F) STEM analysis identified five distinct and significant temporal expression clusters of genes. The median RPKM of each cluster over time is plotted. The Bonferroni-corrected P values for each cluster in order are 2.83e-24, 1.05e-6, 1.10e-20, 9.70e-17, and 1.30e-4. Statistical significance between time points was determined using one-way ANOVA with Dunnett’s multiple-comparison test relative to N (naive). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.