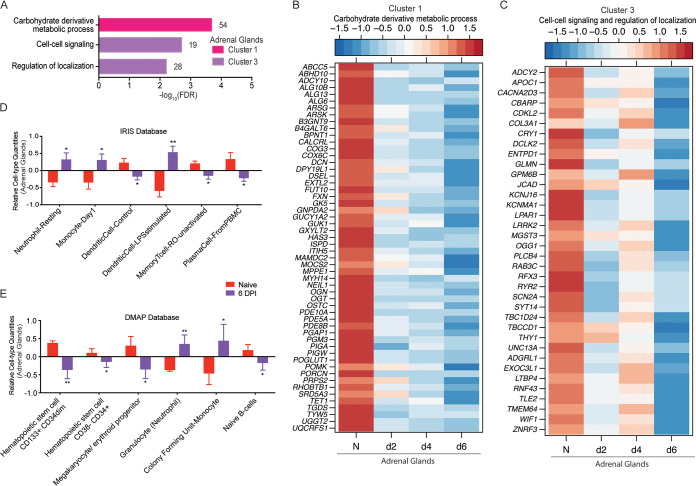
FIG 5.
Adrenal-specific DEGs play a role in metabolic processes and signaling. (A) Functional enrichment of adrenal-specific DEGs within clusters 1 and 3 carried out using MetaCore. The bar graph indicates the –log10 FDR-corrected P value. Numbers next to the bars indicate the number of genes that mapped to each of the GO terms. (B and C) Heatmap of genes in cluster 1 that enriched to “carbohydrate derivative metabolic process” (B) and genes in cluster 3 that enriched to “cell to cell signaling” and “regulation of localization” (C). Each column represents the median normalized transcript counts (RPKM) for each gene at each time point: N (naive; n = 4), d2 (2 DPI; n = 4); d4 (4 DPI; n = 2); d6 (6 DPI; n = 4). The range of colors is based on scaled and centered RPKM values of the entire set of genes, with red indicating highly expressed genes and blue indicating lowly expressed genes. (D to E) Bar graph depicting the mean ± SD of immune-cell-type frequencies predicted using ImmQuant software and the IRIS (D) and DMAP (E) databases in naive samples (n = 4) and those obtained 6 DPI (n = 4). Significance between naive and 6 DPI samples was determined using unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001.