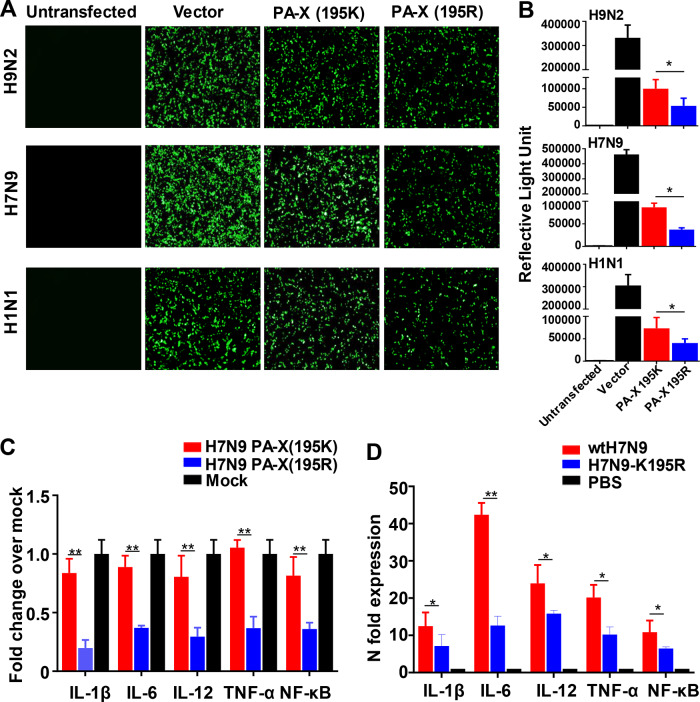
FIG 5.
Effects of PA-X substitutions in suppressing nonviral protein expression and cytokine responses. (A) GFP expression measured at 24 h posttransfection. 293T cells were cotransfected with expression vectors encoding GFP, together with expression vectors carrying either the PA-X (195K) or PA-X (195R) gene of the indicated virus. Untransfected 293T cells or 293T cells cotransfected with GFP expression plasmids and the empty pCAGGS vector served as controls. (B) Luciferase expression from a pRL-SV40 vector encoding Renilla luciferase in 293T cells cotransfected with either empty pCAGGS plasmid (Vector) or pCAGGS encoding the indicated genes after 24 h. The results are shown as means and standard deviations (n = 3 biological replicates; n = 3 technical replicates). (C) Effect of H7N9 PA-X (195K) or H7N9 PA-X (195R) on cytokine mRNA abundance in A549 cells. Total RNA was extracted from transfected cells at 24 h posttransfection, and the abundances of IL-1β, IL-6, IL-12, TNF-α, and NF-κB mRNAs were quantified by qRT-PCR. (D) Effect of PA-X on cytokine mRNA abundance in A549 cells. A549 cells were infected with wtH7N9 virus or H7N9-K195R virus at an MOI of 1. Total RNA was extracted from infected cells at 12 hpi, and the abundances of IL-1β, IL-6, IL-12, TNF-α, and NF-κB mRNAs were quantitated by qRT-PCR. The data are presented as the means and standard deviations of the results of three independent experiments. Statistical significance was assessed using two-way ANOVA (*, P < 0.05; **, P < 0.01).