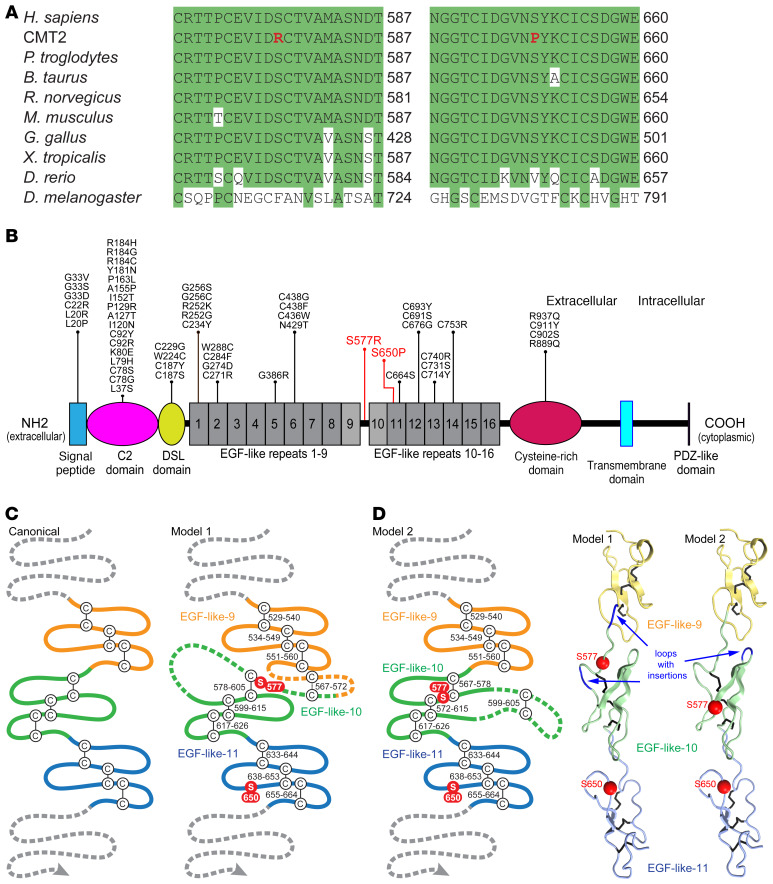
Figure 2. Neuropathy-associated mutations affect conserved amino acids in the extracellular domain of JAG1.
(A) Clustal Omega alignments of the JAG1 protein from divergent species. The p.Ser577Arg and p.Ser650Pro mutations both disrupt serine residues located in highly conserved regions of JAG1. (B) The domain structure of JAG1, with the CMT2-associated mutations indicated in red. Though Alagille syndrome is primarily caused by JAG1 gene deletion or truncating mutations, a number of missense mutations (shown in black) have been described (see Supplemental Methods for references). Of the 42 amino acids mutated in Alagille syndrome, 20 are cysteine residues and none are serine residues; few occur in proximity to Ser577 and Ser650. (C) A schematic of 3 canonical EGF-like repeats is shown, along with 2 proposed models for JAG1 EGF-like repeats 9–11. In model 1, residues 562–585 are split between EGF-like repeats 9 and 10, including a potential disulfide bridge between residues 567 and 572 preceding the start of repeat 10. Model 1 requires 2 insertions, 1 before and 1 within repeat 10. In model 2, residues 562 to 585 are included as a single insertion in the second loop of EGF-like repeat 10. (D) 3D modeling shows that Ser577 and Ser650 may reside on the same surface of the JAG1 extracellular domain (model 2).