Figure 7.
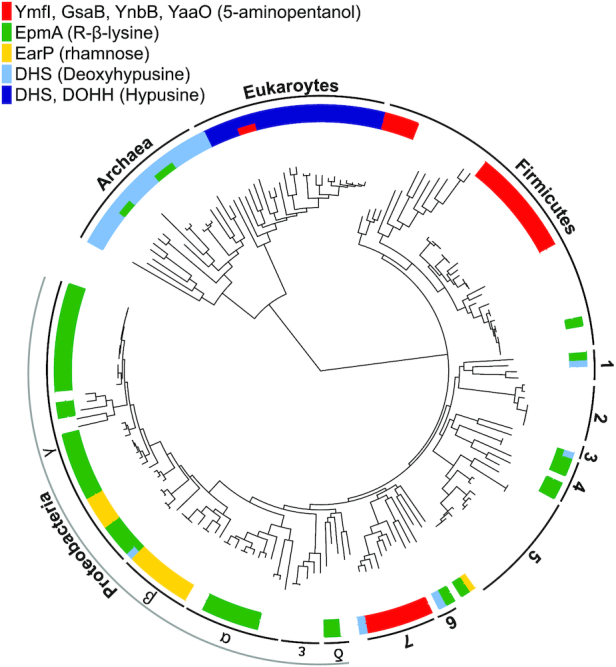
Distribution of the EF-P post-translational modification machinery. Presence of homologs to the EF-P post-translational modification enzymes that have been identified to date across the tree of life. Numbers indicate the following clades (1) Flavobacterium-Cytophaga-Bacteroides group, (2) Chlamydiales, (3) Planctomycetes, (4) Spirochaetes, (5) Actinobacteria, (6) Deinococcus-Thermus group and (7) Cyanobacteria. DHS, EarP and EpmA homologs, were identified as reported in Hummels et al. 2017. DOHH, YmfI, YaaO, GsaB and YnbB homologs were identified using a BLAST search (BLAST + version 2.6.0, Camacho et al. 2009) with an E-value threshold of 1e-20. For each group of enzymes, the associated post-translational modification moiety is listed in parentheses. Where multiple enzyme groups are encoded in the same genome, the bar is split accordingly. White space indicates the absence of homologs to any known sets of EF-P modification enzymes.
